Proteins serve as the carriers of life and the executors of biological functions, playing a crucial role in maintaining normal physiological activities, including cytoskeleton construction, enzymatic catalysis, signal transduction, molecular transport, and immune system regulation.
The function of a protein is closely related to its specific structure. Therefore, understanding the structure and interactions of proteins is crucial to comprehending how organisms work. Protein characterization is the key step to studying protein structure and function.
Contents
1. What Is Protein Characterization?
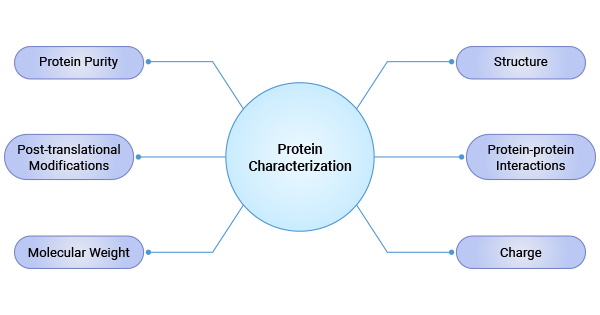
Protein characterization refers to the process of determining and describing the structure, properties, and biological function of proteins. Protein characterization analysis involves assessing protein purity, molecular weight, structure, post-translational modifications, and interactions, among other factors.
Figure 1. Protein Characterization
2. Why Is Protein Characterization Important?
Protein characterization is central to bio-pharmaceutical research, revealing crucial details about a protein's structure, function, and interactions, which are vital for investigating pathogenesis, developing drugs, and ensuring the safety and efficacy of therapeutic proteins.
Protein characterization is important in multiple areas:
Unveiling the mechanisms of biological and pathological processes: Understanding the structure and interactions of proteins can help to grasp the fundamental principles behind biological processes, such as cell signaling, gene expression regulation, and disease pathogenesis.
Drug discovery and development: Protein characterization is crucial for formulating strategies for drug discovery and development. Understanding how drugs interact with proteins can design more effective medications and improve the selectivity and specificity of drugs while minimizing side effects.
Biotechnology and bioengineering: Understanding the structure and function of proteins can guide the design and engineering of recombinant proteins to meet the specific needs of various applications.
3. How Are Proteins Characterized?
Proteins are characterized using various techniques to determine their composition, structure, function, interactions, and other properties.
| Protein Characterization Methods |
Mechanism |
Application |
Advantages |
Disadvantages |
| Mass Spectrometry (MS) |
Ionization of the protein, separation and detection of the ion, determination of the mass-to-charge ratio of charged ions, and final comparison with the mass spectrum database [1] |
Protein sequencing, detecting PTMs |
High sensitivity and specificity, high resolution;
Can analyse complex sample mixtures |
Samples need to be pre-treated;
Requires specialized equipment and expertise |
| SDS-PAGE |
Separation of proteins based on size and charge using an electric field applied to a gel matrix [2,3] |
Isolation of proteins according to their molecular size and charge |
Simple and widely used technique |
Limited resolution for large proteins;
Can not provide structural information |
| Western blot (WB) |
Detection of specific proteins using antibodies for identification and quantification[4,5] |
Isolation and identification of individual proteins |
High specificity and sensitivity |
Labor-intensive and time-consuming process |
| Circular Dichroism (CD) Spectroscopy |
Measurement of the differential absorption of left- and right-handed circularly polarized light by chiral molecules to analyze protein secondary structure [6,7] |
UV CD: determine protein secondary structure;
IR CD: study the structure of small organic molecules, proteins, and DNA;
UV/Vis CD: investigate charge transfer transitions in metal-protein complexes
|
Provides information on protein secondary structure;
Can be used for rapid analysis |
Limited resolution for complex proteins |
| X-ray Crystallography |
Analysis of protein crystal diffraction patterns to determine atomic-level structure [8,9] |
Elucidation of the detailed 3-dimensional structure of proteins |
High resolution and precision;
Can provide detailed protein structure information |
Requires protein crystallization;
Analysis o the structure takes a lot of time and resources |
| Nuclear Magnetic Resonance (NMR) Spectroscopy |
Detection of signals from atomic nuclei to determine protein structure in solution [10] |
|
High resolution and non-destructive;
Provides atomic-level structural and dynamic information |
Difficult for large molecule samples, and require a high concentration of samples;
Requires isotopic labeling and may be time-consuming |
| Surface Plasmon Resonance (SPR) |
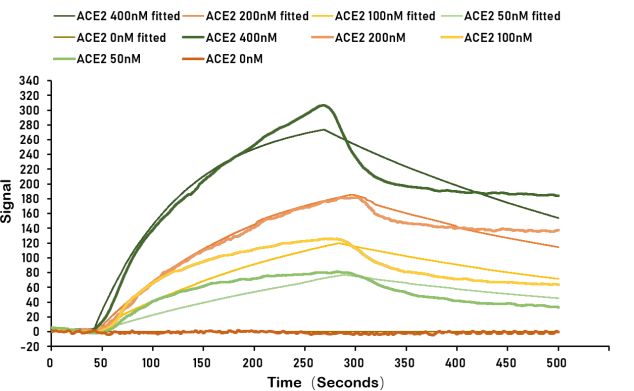
Detection of changes in refractive index upon binding of proteins to metal surface plasmas [11,12] |
|
Detection of protein interactions in real time;
High sensitivity |
Expensive equipment and consumables; Limited to studying interactions with surface-immobilized ligands |
Mass spectrometry, X-ray crystallography, NMR, and SPR are common methods used for protein characterization. Each method has its own advantages and limitations. Choosing the optimal technique to analyze a protein sample requires considering factors such as sample properties, research objectives, and experimental conditions.
Mass spectrometry is commonly used for proteins with low molecular weight. X-ray crystallography and NMR can provide more detailed structural information for proteins with high molecular weight. SPR is suitable for studying the dynamic changes and interaction networks of proteins. Therefore, when selecting a protein characterization method, it's essential to consider these factors comprehensively and choose accordingly based on actual needs.
4. Applications of Protein Characterization
Protein characterization is essential in various fields and applications. Here are some key areas where protein characterization methods are widely used:
4.1 Laboratory Research
Protein characterization in laboratory research helps to study biological processes, modify proteins for improved or novel functions in protein engineering, and provide fundamental insights into molecular biology and biochemistry.
4.2 Biomedical Research
Characterizing proteins involved in diseases helps understand their roles and mechanisms. Protein characterization also contributes to discovering biomarkers for specific diseases, developing personalized treatment strategies, or identifying potential targets for novel drugs.
4.3 Pharmaceutical Industry
Protein characterization is essential in the pharmaceutic industry, including designing and optimizing drugs that target specific proteins, ensuring the purity, safety, and efficacy of therapeutic proteins, and studying how drugs interact with proteins in the body.
4.4 Food Industry
Protein characterization is important for nutritional analysis, food safety, and quality control in the food industry. Characterizing food products can determine the protein content, detect allergens, and ensure the safety of the food products. Protein characterization guides the formulation of food products, ensuring they meet consumer expectations for quality, functionality, and nutritional value.
These applications highlight the versatility and importance of protein characterization in advancing science and technology across multiple disciplines.
5. Examples of Protein Characterization
CUSABIO can provide some recombinant proteins that have been well characterized.
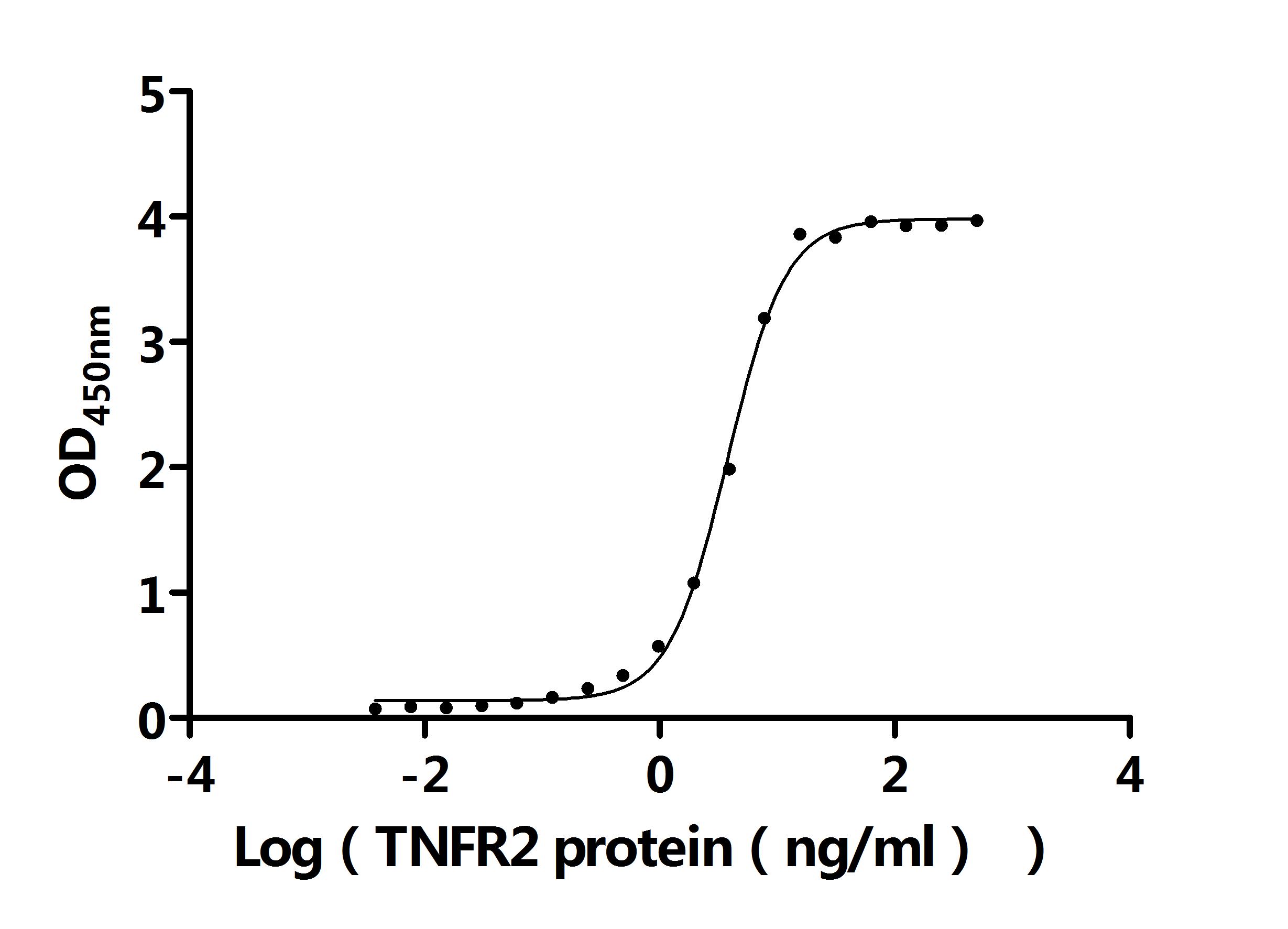
● Recombinant Human CD276 antigen (CD276), partial (Active)
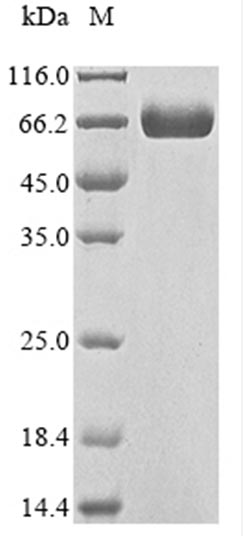
Recombinant human CD276 antigen's molecular weight was about 60 kDa measured by SDS-PAGE.
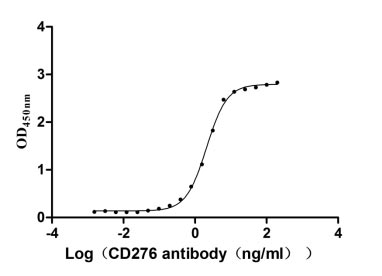
Recombinant human CD276 antigen's activity was measured by its binding ability in a functional ELISA. Immobilized CD276 at 2 μg/ml can bind the anti-CD276 rabbit monoclonal antibody, the EC50 of human CD276 protein is 1.961-2.243 ng/ml.
Conclusion
Protein characterization plays a vital role in understanding protein structure, function, and interactions. By characterizing proteins, researchers can determine their composition, sequence, folding patterns, and post-translational modifications, providing insights into their biological functions. Additionally, protein characterization helps identify protein-protein interactions, enabling the study of complex biological processes and pathways, disease mechanisms, and drug discovery.
References
[1] Zhang, G., Ueberheide, B. M., et al. (2010). Protein Quantitation Using Mass Spectrometry [J]. Methods in Molecular Biology (Clifton, N.J.), 673, 211.
[2] Nowakowski, A. B., Wobig, et al. (2014). Native SDS-PAGE: High Resolution Electrophoretic Separation of Proteins With Retention of Native Properties Including Bound Metal Ions [J]. Metallomics : Integrated Biometal Science, 6(5), 1068.
[3] Hagiwara, M. (2022). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blotting analyses via colored stacking gels [J]. Analytical Biochemistry, 652, 114751.
[4] W.N. Burnette. Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A [J]. Anal. Biochem., 112 (1981), pp. 195-203.
[5] C.P. Moritz. 40years Western blotting: a scientific birthday toast [J]. J. Proteonomics, 212 (2020), Article 103575.
[6] Greenfield, N. J. (2006). Using circular dichroism spectra to estimate protein secondary structure [J]. Nature Protocols, 1(6), 2876.
[7] Greenfield, N.J. (2004). Circular Dichroism Analysis for Protein-Protein Interactions. In: Fu, H. (eds) Protein-Protein Interactions [J]. Methods in Molecular Biology, vol 261. Humana Press.
[8] Gawas, U. B., Mandrekar, V. K., & Majik, M. S. (2018). Structural analysis of proteins using X-ray diffraction technique [J]. Advances in Biological Science Research, 69-84.
[9] Maveyraud, L., & Mourey, L. (2020). Protein X-ray Crystallography and Drug Discovery [J]. Molecules, 25(5).
[10] Hu Y, Cheng K, He L, et al. NMR-Based Methods for Protein Analysis [J]. Anal Chem. 2021 Feb 2;93(4):1866-1879.
[11] Drescher, D.G., Drescher, M.J. (2023). Protein Interaction Analysis by Surface Plasmon Resonance. In: Sousa, Â., Passarinha, L. (eds) Advanced Methods in Structural Biology [J]. Methods in Molecular Biology, vol 2652. Humana, New York, NY.
[12] Douzi B. Protein-Protein Interactions: Surface Plasmon Resonance [J]. Methods Mol Biol. 2017;1615:257-275.
CUSABIO team. What Is Protein Characterization: Definition, Examples, Methods, and Techniques. https://www.cusabio.com/c-21178.html






Comments
Leave a Comment