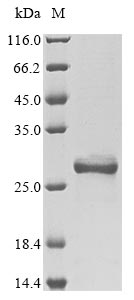
Recombinant Mouse High mobility group protein B1 (Hmgb1) is produced in E. coli and contains the amino acid sequence from 2 to 215. The protein is partially expressed with an N-terminal 6xHis-tag to make purification easier. SDS-PAGE analysis confirms it reaches a purity level of greater than 85%. This product is for research use only and should not be used in clinical applications.
High mobility group protein B1 (Hmgb1) is a non-histone chromosomal protein that appears to play a critical role in DNA architecture and regulation. The protein is involved in various cellular processes, including DNA repair, transcription, and replication. How Hmgb1 interacts with DNA and other nuclear proteins makes it an important subject when studying chromatin dynamics and gene expression.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
Based on the provided information, the recombinant mouse HMGB1 protein may not be correctly folded or fully bioactive without experimental validation. HMGB1 is a DNA-binding protein that requires proper folding of its two HMG boxes (A and B domains) for function. While E. coli expression systems can produce soluble HMGB1, the N-terminal 6xHis tag may sterically hinder the DNA-binding domains, as the DNA-binding sites are located near the N-terminus. The protein is partial length (2-215aa), which is nearly full-length but lacks the first amino acid; this might not significantly impact folding, but the tag could. The purity of >85% indicates the presence of impurities, including potential misfolded proteins or aggregates. Without validation (e.g., circular dichroism for secondary structure, DNA-binding assays like EMSA), the folding status and bioactivity remain uncertain.
1. Protein-DNA Interaction Studies
This recombinant mouse HMGB1 protein can be used to investigate DNA-binding properties only if correct folding and DNA-binding activity are experimentally verified. The N-terminal His-tag may interfere with DNA binding, leading to inaccurate results in SPR or EMSA assays. If misfolded, binding specificity studies may yield false negatives or altered kinetics. It is recommended to validate folding and DNA binding with positive controls (e.g., tag-free HMGB1) before use. The mouse origin is suitable for species-specific studies but does not guarantee functionality.
2. Antibody Development and Validation
This purified recombinant HMGB1 can serve as an antigen for antibody generation, but antibody specificity depends on the protein's folding state. If correctly folded, antibodies may recognize native epitopes; if misfolded, they could target non-conformational epitopes, reducing their utility for detecting physiological HMGB1. The His-tag may dominate the immune response, leading to tag-specific antibodies. For reliable outcomes, validate antibodies against full-length, tag-free HMGB1 or endogenous protein. The purity of>85% is acceptable but risks cross-reactivity with contaminants.
3. Protein-Protein Interaction Analysis
This HMGB1 protein can be used in pull-down assays only if folding is validated, as misfolding may cause non-specific interactions or false negatives. The partial length (2-215aa) retains major domains but may lack full interaction capabilities if the C-terminal region is critical. The His-tag allows immobilization but could promote tag-mediated artifacts. Include controls (e.g., tag-only protein) and verify folding with biophysical methods before interaction studies.
4. Structural and Biophysical Characterization
This recombinant HMGB1 is unsuitable for high-resolution structural studies (e.g., crystallography or NMR) without tag removal, as the His-tag introduces heterogeneity and flexibility. Biophysical techniques (e.g., CD, DLS) can assess general folding but may be confounded by the tag. The purity >85% is suboptimal for structural work (typically requiring >95%). For meaningful insights, remove the tag and confirm native-like structure through DNA-binding assays first.
5. In Vitro Functional Assays
This HMGB1 protein can be used in chromatin remodeling assays only if bioactivity is confirmed. The His-tag may affect DNA bending or nucleosome positioning, leading to misleading functional data. If misfolded, results may not reflect physiological HMGB1 behavior. Validate activity with known assays (e.g., nucleosome sliding) and compare with tag-free standards. The mouse sequence is appropriate for mouse-derived systems but requires folding validation.
Final Recommendation & Action Plan
To ensure reliable outcomes, first validate the folding and bioactivity of the recombinant HMGB1 using techniques such as circular dichroism to confirm expected secondary structure (e.g., alpha-helical content in HMG boxes), electrophoretic mobility shift assays (EMSA) to verify DNA-binding capability, and size-exclusion chromatography to assess oligomeric state and purity. If possible, remove the 6xHis tag via proteolytic cleavage and re-purify the tag-free protein for functional and structural studies. For applications like antibody development, proceed with caution and validate antibodies thoroughly. Always include appropriate controls, such as tag-free HMGB1 or DNA-binding mutants, to account for potential artifacts. Given the E. coli expression system, while it can produce functional HMGB1, independent validation is critical due to the tag and purity concerns.