p53 is a tumor suppressor gene that is involved in the development of most cancers in humans. The p53 protein is extremely important for inhibiting the development of cancer. Studies have shown that in addition to tumor suppressor function, p53 is also involved in a variety of physiological processes such as apoptosis, DNA damage repair, regulation of body metabolism, and senescence. Recent studies have found that p53 is also involved in the regulation of antiviral immune responses in the body.
1. The Characteristics of p53
Tumorigenesis is a multi-gene, multi-stage process. A large number of studies in recent years have shown that p53 missense mutation or allele deletion has a high incidence in many tumors [1]. According to statistics, in all malignant tumors, the mutation probability of this gene is more than 50%. Currently, p53 gene has the highest correlation with liver cancer, breast cancer, ovarian cancer, brain tumor, gastric cancer, bowel cancer, esophageal cancer, lung cancer, bladder cancer, leukemia and other human tumors.
Since its first discovery in 1979, the p53 gene has become an important part of current molecular biology research [2].
The p53 gene is a very important tumor suppressor gene in the human body. It can inhibit the progression of the cell cycle, inhibit cell proliferation, and prevent tumorigenesis by causing apoptosis in damaged cells.
p53 can be divided into wild type and mutant type. Wild-type (wt-p53) protein is an important transcription factor, which blocks cell cycle, induces apoptosis and senescence, participates in DNA damage repair, and inhibits angiogenesis by regulating downstream target genes (p21, Bax, PTEN, p48, PAI, etc.) to prevent the occurrence and development of tumor. The wild-type p53 protein has a short half-life and low content in cells, so it is difficult to detect by immunohistochemistry, while the mutant p53 protein has a high content in cells and a long half-life, which can be detected by immunohistochemistry. Mutant p53 protein promotes the development of cancer.
2. Structure of p53 Protein
p53 gene, located on the short arm of human chromosome 17, is about 20kb in length and contains 11 exons and 10 introns. Its mRNA length is 2500bp. It consists of 393 amino acids with a molecular weight of 53kD. p53 protein can be detected in almost all somatic cells.
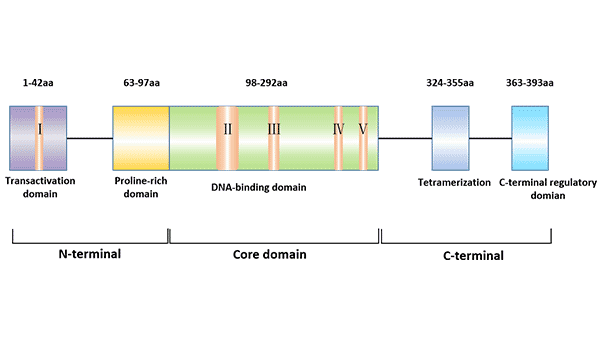
Figure 1 The structure of p53 protein
There are 5 highly conserved regions of p53, which are key loci for p53 function [3]. The main functional regions of p53 protein include: transcriptional activation region at the N-terminal (1 ~ 42aa), DNA binding region at the intermediate site (102 ~ 292aa), polyaggregation region (323 ~ 356aa) and regulatory region (360 ~ 393aa) at the C-terminal.
-
The N-terminal region of p53 plays a role in recruiting transcription proteins. Adenovirus E1B protein, human MDM2, and hepatitis b virus X protein can bind to the N-terminus of p53 and inhibit its transcriptional activation function. The N-terminus also has a proline-rich region and may be involved in apoptosis regulated by p53.
-
The intermediate region of p53 protein (102 ~ 292aa) contains four conserved regions, where 80% ~ 90% of the mutations caused by tumor cells occur.
-
The C-terminus of p53 (300-393aa) contains a variable junction region that connects the central core region to the C-terminal region; there is also a tetramerization region. There are also three nuclear localization signals (NLS) at the C-terminus of p53: NLS1 (316-325aa), NLS2 (369-375aa), and NLS3 (379-384aa). The C-terminus is a flexible unfolded region that binds to DNA non-specifically and acts as a repressor in the regulation of other genes.
The spatial structure of p53 protein determines its biological function.
3. p53 Protein Function
The regulation mechanism and function of p53 protein are mainly characterized by cell cycle regulation, DNA damage repair, apoptosis, cell senescence, regulation of metabolism.
Factors regulating p53 protein expression: IFN-α/IFN-β can be involved in the regulation of p53 mRNA transcription and protein expression, and this process requires the involvement of IRF9 [4].
In human dermal fibroblasts, Gld2 regulates the expression of cytoplasmic polyadenylation element binding protein (CPEB) by regulating the stability of miR-122, whereas CPEB can The 3'-UTR binding of the p53 gene then recruits Gld4 to maintain p53 mRNA polyadenylation and transcription [5].
3.1 p53 and Cell Cycle Regulation
In addition to being in the form of p53 protein, p53 is present in cells as a potential untranscribed form. The untranscribed form of p53 is only activated to produce biological functions. p53 protein can be activated by DNA damage. The upstream factor of p53 recognizes DNA-damaged proteins.
In addition, p53 can also bind directly to the ends and damage sites of DNA, thereby being phosphorylated or receiving other signals to generate activity. In addition to DNA damage, tissue hypoxia also increases and activates p53 concentration.
p53-mediated downstream processes mainly include:
-
Make the cells stop at the cell cycle test point
-
Make the cells go to apoptosis
p53 gene has the function of blocking cell cycle. The expression or ectopic expression of p53 after DNA damage keeps the cell cycle at the G1/S test point.
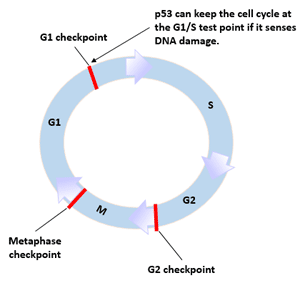
Figure 2 p53 and cell cycle
3.2 p53 and DNA Damage Repair
The p53-regulated DNA damage repair process mainly includes Nucleotide Excision repair (NER), Base excision repair (BER), Nonhomologous end-joining (NHEJ) and transcriptional activation pathways and Homologous recombination by both transactivation-dependent and-independent pathways [6].
3.3 p53 and Apoptosis
p53 gene has the effect of promoting cell apoptosis. p53 protein can induce the apoptosis of malignant tumor cells, thus damaging the function of tumor cells and eventually causing tumor atrophy or disappearance. It can regulate apoptosis through proteins such as Bax/Bcl-2, Fas/Apo1, and induce apoptosis through death signal receptor proteins such as TNF receptor and Fas protein. p53 can also directly stimulate mitochondria to release highly toxic oxygen free radicals to induce apoptosis [7].
The role of p53 in promoting apoptosis is closely related to its tumor suppressor function. Apoptotic mechanisms regulated by p53 include transcriptional activation and non-transcriptional activation pathways [8].
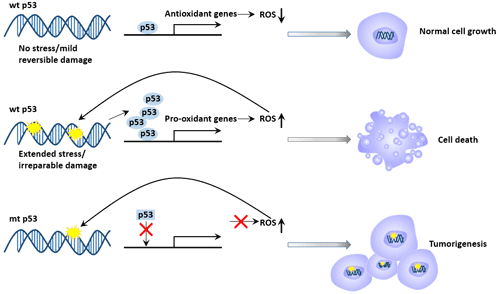
Figure 3 p53 and DNA damage repair and apoptosis
3.4 p53 and Cellular Senescence
Cell senescence can inhibit the occurrence of cell carcinogenesis, which is mainly regulated by two pathways, p53 and p16/Rb. Cell senescence induced by p53 pathway can be activated by RAS mitotic signaling such as telomerase inactivation and reactive oxygen stimulation.
When telomeres are shortened to produce cell replicative senescence, p53 can detect telomere shortening [9] and signal telomere dysfunction to ATM protein kinase. The specific mechanism of p53 to regulate cell senescence is still not clear, but some studies have shown that the specific mechanism can be activated through p21 [10].
3.5 p53 and Metabolic Regulation
The metabolic regulation of p53 on the body is mainly manifested in three aspects:
-
p53 regulates the production of energy in the body. p53 lowers the glycolytic pathway (through the glucose transporters GLUT1, GLUT4, and NF-κB), increases transcription of cytochrome oxidase subunit I and activates cytochrome oxidase 2 (SCO2) synthesis.
-
p53 regulates cell growth. In the case of cell nutritional deficiency, p53 can regulate the AMPK pathway by activating the expression of AMPK and TSC2 genes, thus playing an important regulatory role in the process of cell proliferation and growth.
-
p53 regulates autophagy. Damage-regulated autophagy modulator (DRAM), which regulates autophagy, may be a target gene of p53, and some target genes of p53, such as BAX and PUMA, may participate in autophagy [11].
3.6 The Antiviral Effect of p53
Tumor suppressor p53 plays an important role in tumor suppression due to its regulation of cell cycle and the promotion of apoptosis to prevent the emergence of mutant cells. p53 can participate in the apoptosis caused by the virus, thereby inhibiting the replication and spread of the virus. p53 can participate in innate immunity, acquired immunity and inflammatory response to play an antiviral role.
4. The Relationship between p53 and Related Genes
4.1 p53 and Mdm2
Mdm2 is an evolutionarily conserved gene with transcription factor function. It binds to wild-type (wt) or mutant (mt) p53, which inhibits p53 function, thereby arresting the G1 phase of the cell and re-entering the cell cycle. It is believed that p53 protein binds to mdm2 protein to form oligodimer complex, and overexpressed mdm2 inhibits p53 transcriptional activity; wild-type p53 expression in turn affects mdm2 protein expression in the same cell. There is a mutual regulation mechanism in regulating cell division and proliferation.
4.2 p53 and p21WAF1/CIPI
The p21WAF1/CIP1 gene encodes a cyclin-dependent kinase inhibitory protein (CDKI) that is involved in cell growth, differentiation, senescence, and death by regulating cell cycle progression. As a direct downstream target gene of p53, p21WAF1 is a key gene involved in cell cycle regulation regulated by p53, and can perform part of the function of p53.
Both p53 and p21 are involved in cellular DNA damage repair. When the DNA of the cell is damaged, the p53 protein accumulates, and the expression of p21 gene is up-regulated, and the cell is arrested in the G1 phase. Mutual regulation of p53 and p21 is necessary to arrest damaged cells in both G1 and G2 phases.
5. The Anticancer Mechanism of p53 Gene
The occurrence of tumors is mainly caused by the disorder of cell cycle regulation. Apoptosis and senescence are two tumor suppressor pathways in cells, and the p53 gene plays an important role in both aspects.
Wild-type p53 protein can bind to DNA of specific sequences, activate the transcriptional activity of gene promoters containing p53 response elements, inhibit the transcriptional activity of gene promoters without p53 response elements [12], and inhibit DNA replication. Wild-type p53 prevents cells from remaining in the G1 or G0 phase, allowing enough time for damaged DNA to be repaired. If it cannot be repaired, programmed death will occur in cells with DNA damage [13].
The function of p53 is affected by many factors, including cell differentiation, cell senescence, DNA damage and hypoxia. The activation of p53 by cells mainly increases the expression and stability of p53, changes the cell localization of p53, and increases its biological activity.
The stability of p53 is mainly increased by the action of MDM2, which can inhibit the transcriptional activity of p53 and promote the degradation of p53. ARF binds directly to MDM2 and blocks its binding to p53 to prevent degradation of p53 [14].
Intracellular localization of p53: When stimulated, p53 enters the nucleus via microtubules and dynein, a process that relies primarily on its C-terminal nuclear import sequence [15]. When p53 is to be degraded, it needs to be transported out of the nucleus for effective degradation by combining MDM2 with its c-terminal nuclear output sequence [16] [17].
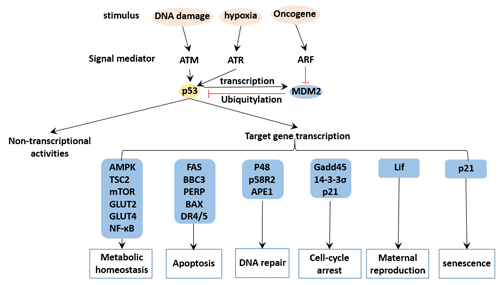
Figure 4 p53 regulation network
6. Application of p53
6.1 Application of p53 in Gene Therapy
The important role of p53 in tumor suppression makes p53 and its complexes a target for the development of new anticancer drugs.
As for the application of p53 gene in tumor gene therapy, adenovirus is the most commonly used vector to introduce normal wild-type p53 gene into tumor cells to replace the mutant p53 gene. A large number of studies have shown that adenovirus-mediated p53 gene infection can effectively control tumor cell growth or induce apoptosis. In addition, non-viral vector liposomes are also considered as a potential therapeutic vector.
Currently, the main therapeutic strategies include inhibition of mdm2-p53 interaction, degradation of mutant p53 protein, inhibition of mutant p53 downstream pathway, RNA interference of mutant p53 expression, and restoration of mutant p53 conformation.
6.2 Application of p53 Gene in Cancer
-
Ultra-early early warning detection of tumor
Tumors are the result of the accumulation of genetic mutations. During the onset and evolution of tumors, the human body has almost no symptoms. Early detection of tumors through correlation mutation detection and reasonable intervention can reverse the formation of tumors.
-
Monitoring tumor recurrence after surgery
Scanning for p53 mutations in tumor tissues can monitor postoperative recurrence and assess efficacy.
6.2.1 Detection of p53 Gene Expression
The monitoring of p53 gene provides a basis for the early diagnosis and prediction of tumors, and can make scientific inference on the occurrence, development and prognosis of tumors. The detection methods mainly include:
-
Nano-Probe Detection
Gold nanoparticles have the advantages of surface effect, small size effect and better biocompatibility [18].
-
Immunohistochemistry
The principle of this method is to use the specific antibody to react with the p53 protein to develop an antigen-antibody color reaction, and indirectly determine the presence or absence of p53 protein according to the presence or absence of the color spot. This method has been widely used, but it is not possible to screen samples in large quantities.
-
Isotope Immunoassay
Use isotope labeling for antigen antibody reaction to detect proteins. This method has the advantages of high accuracy, small sample consumption, easy standardization and automation [19].
-
Enzyme-Linked Immunosorbent Assay
This method has strong sensitivity, specificity, repeatability and stability [20]. However, this method is prone to false positives and cumbersome operation.
In addition, flow cytometry assay and tissue microarray assay can be used for the detection of p53 protein.
References
[1] Pellegata N S, Ranzani G N. The significance of p53 mutations in human cancers [J]. European Journal of Histochemistry Ejh, 1996, 40(4): 273.
[2] Greenblatt M. Mutatinons in the p53 tumor suppressor gene. Clues to cancer etiology and molecular pathogenesis [J]. Cancer Res, 1994, 54.
[3] Soussi T, May P. Structural Aspects of the p53 Protein in Relation to Gene Evolution: A Second Look [J]. Journal of Molecular Biology, 1996, 260(5): 0-637.
[4] Wang B, Xiao Z, Ren E C. Redefining the p53 response element [J]. Proceedings of the National Academy of Sciences, 2009, 106(34): 14373-14378.
[5] Burns D M, D’Ambrogio, Andrea, et al. CPEB and two poly (A) polymerases control miR-122 stability and p53 mRNA translation [J]. NATURE, 2011, 473(7345): 105-108.
[6] Adimoolam S, Ford J M. p53 and regulation of DNA damage recognition during nucleotide excision repair [J]. DNA Repair, 2003, 2(9): 0-954.
[7] Oda E. Noxa, a BH3-Only Member of the Bcl-2 Family and Candidate Mediator of p53-Induced Apoptosis [J]. Science, 2000, 288(5468): 1053-1058.
[8] Yee, K. S. Complicating the complexity of p53 [J]. Carcinogenesis, 2005, 26(8): 1317-1322.
[9] Lansdorp P M. Repair of telomeric DNA prior to replicative senescence [J]. Mechanisms of Ageing & Development, 2000, 118(1): 23-34.
[10] Itahana K, Dimri G, Campisi J. Regulation of cellular senescence by p53 [J]. Febs Journal, 2010, 268(10): 2784-2791.
[11] Wilkinson S, James J, Ryan K M, et al. PUMA- and Bax-induced autophagy contributes to apoptosis [J]. Cell Death & Differentiation, 2009, 16(8): 1135-1145.
[12] Fields S, Jang S. Presence of a potent transcription activating sequence in the p53 protein [J]. Science, 1990, 249(4972): 1046-1049.
[13]Lane D. P. A death in the life of p53 [J]. Nature, 1993, 362(6423): 786-787.
[14] Hirao A. DNA Damage-Induced Activation of p53 by the Checkpoint Kinase Chk2 [J]. Science, 2000, 287(5459): 1824-1827.
[15] Kevin M Ryan, Andrew C Phillips, Karen H Vousden. Regulation and function of the p53 tumor suppressor protein [J]. Current Opinion in Cell Biology, 2001, 13(3): 332-337.
[16] Boyd S D, Tsai K Y, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53 [J]. Nature Cell Biology, 2000, 2(9): 563–568.
[17] Geyer R, Yu K N, Maki K C. The MDM2 RING-finger domain is required to promote p53 nuclear export [J]. Nature Cell Biology, 2000, 2(9): 569-573.
[18] Bao Y P, Wei T F, Lefebvre P A, et al. Detection of Protein Analytes via Nanoparticle-Based Bio Bar Code Technology [J]. Analytical Chemistry, 2006, 78(6): 2055-2059.
[19] Cioffi M, Vietri M T, Gazzerro P, et al. Serum anti-p53 antibodies in lung cancer: comparison with established tumor markers [J]. Lung Cancer, 2001, 33(2-3): 163-169.
[20] Shiota G, Kishimoto Y, Suyama A, et al. Prognostic significance of serum anti-p53 antibody in patients with hepatocellular carcinoma [J]. Journal of Hepatology, 1997, 27(4): 661-668.
CUSABIO team. The True Face of Tumor Suppressor Gene p53. https://www.cusabio.com/c-20904.html








Comments
Leave a Comment