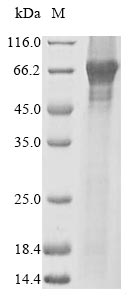
The recombinant Human PPARA was expressed with the amino acid range of 1-468. The expected molecular weight for the PPARA protein is calculated to be 56.2 kDa. This protein is generated in a baculovirus-based system. The N-terminal 10xHis tag and C-terminal Myc tag was fused into the coding gene segment of PPARA, making it easier to detect and purify the PPARA recombinant protein in the later stages of expression and purification.
PPARA (Peroxisome proliferator-activated receptor alpha) is a nuclear receptor playing a crucial role in energy and lipid metabolism. Research primarily focuses on two major areas. Firstly, the role of PPARA in lipid metabolism regulation is widely studied. Researchers delve into the regulatory mechanisms of PPARA in fatty acid oxidation, activation of brown adipose tissue, cholesterol metabolism, providing new insights into the treatment of metabolic diseases like diabetes and hyperlipidemia. Secondly, the physiological functions of PPARA in tissues such as the liver and heart have attracted considerable attention. Researchers are attentive to PPARA's regulatory role in liver lipid metabolism, cardiac protection, and its involvement in metabolism-related inflammatory responses.