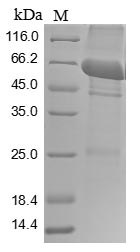
Recombinant Rat Pyruvate Kinase PKM is produced in a baculovirus expression system, covering the full-length protein sequence from amino acids 1 to 531. It features an N-terminal 10xHis-tag and a C-terminal Myc-tag, ensuring efficient purification and detection. The protein is purified to over 85% purity, as verified by SDS-PAGE, and is designed for research use only. This appears to provide a reliable tool for experimental applications.
Pyruvate kinase PKM plays a crucial role in glycolysis, catalyzing the conversion of phosphoenolpyruvate to pyruvate with the generation of ATP. This enzyme is essential for energy production and participates in various metabolic pathways. Its activity and regulation seem critical for understanding metabolic processes, which may make it a valuable target in research focused on cellular energy dynamics and metabolic diseases.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
Rat pyruvate kinase PKM is a tetrameric glycolytic enzyme that requires precise folding, proper oligomerization, and specific allosteric regulation for its catalytic activity. The baculovirus-insect cell expression system provides a eukaryotic environment that supports proper protein folding, post-translational modifications, and complex assembly, significantly increasing the probability of correct tetramer formation compared to prokaryotic systems. However, the dual N-terminal 10xHis-tag and C-terminal Myc-tag may sterically interfere with the protein's oligomerization interfaces or allosteric regulatory sites. While the full-length protein (1-531aa) contains all functional domains, experimental validation remains essential to confirm structural integrity and enzymatic activity.
1. Enzyme Kinetics and Metabolic Pathway Studies
This application is highly dependent on proper folding and tetramerization validation. Pyruvate kinase activity requires precise quaternary structure formation for catalytic function and allosteric regulation. If correctly folded and active (verified through kinetic assays), the protein is suitable for detailed enzymatic studies. If misfolded/inactive (unverified), kinetic measurements will yield biologically meaningless results. The tags may potentially interfere with allosteric effector binding sites.
2. Protein-Protein Interaction Screening
This application carries a significant risk without proper oligomerization validation. PKM interactions with metabolic partners require a correct tetrameric structure. If correctly assembled (verified), the protein can identify physiological interaction partners in glycolytic complexes. If misfolded/misassembled (unverified), there is a high risk of non-specific binding or failure to present genuine interaction interfaces.
3. Antibody Development and Validation
This application is highly suitable regardless of folding status. Antibody development relies on antigenic sequence recognition rather than functional oligomerization. The full-length protein provides comprehensive epitope coverage for generating both linear and conformational antibodies. The high purity ensures minimal contamination-related issues in immunization protocols.
4. Structural and Biophysical Characterization
These studies are essential priority applications for determining the folding and oligomerization status of the protein itself, not the native PKM. Techniques should include size-exclusion chromatography with multi-angle light scattering to verify tetramer formation, circular dichroism spectroscopy to assess secondary structure, and thermal shift assays to evaluate stability. However, the tags may interfere with crystallization for structural studies.
5. Comparative Species Analysis and Evolution Studies
Meaningful comparative studies require native conformation and functional activity. If correctly folded and active (verified), the protein enables valid evolutionary comparisons of kinetic properties and regulatory mechanisms. If misfolded/inactive (unverified), comparative analyses would yield misleading evolutionary insights.
Final Recommendation & Action Plan
The baculovirus expression system provides favorable eukaryotic folding conditions for this tetrameric metabolic enzyme, but the dual-tag configuration and complex oligomerization requirements necessitate rigorous validation before functional applications. Begin with Application 4 (Structural and Biophysical Characterization) to assess oligomerization state through SEC-MALS and validate enzymatic activity using standard pyruvate kinase assays. Once correct tetramer formation and functional activity are verified, proceed cautiously with Applications 1, 2, and 5 for kinetic studies, interaction screening, and comparative analyses. Application 3 (antibody development) can proceed immediately. Always include appropriate controls: validate key findings with tag-free constructs when possible, use known allosteric effectors as positive controls, and consider the tags' potential interference in data interpretation. For critical structural studies requiring high-resolution data, consider using tag-free protein constructs.