| Image |
-
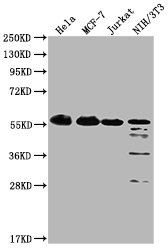
Western Blot
Positive WB detected in: Hela whole cell lysate, MCF-7 whole cell lysate, Jurkat whole cell lysate, NIH/3T3 whole cell lysate
All lanes: PKM antibody at 1:4000
Secondary
Goat polyclonal to Mouse IgG at 1/10000 dilution
Predicted band size: 58 kDa
Observed band size: 58 KDa
Exposure time: 1min
-
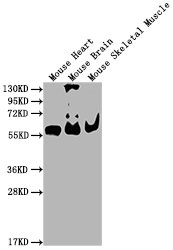
Western Blot
Positive WB detected in: Mouse Heart tissue, Mouse Brain tissue, Mouse Skeletal Muscle tissue
All lanes: PKM antibody at 1:4000
Secondary
Goat polyclonal to Mouse IgG at 1/10000 dilution
Predicted band size: 58 kDa
Observed band size: 58 KDa
-
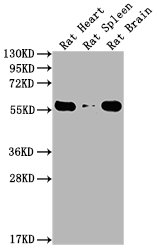
Western Blot
Positive WB detected in: Rat Heart tissue, Rat Spleen tissue, Rat Brain tissue
All lanes: PKM antibody at 1:4000
Secondary
Goat polyclonal to Mouse IgG at 1/10000 dilution
Predicted band size: 55-60 kDa
Observed band size: 55-60 kDa
-
Western Blot
Positive WB detected in: MCF-7 whole cell lysate at 40µg, 20µg, 10µg, 5µg, 2.5µg, 1.25µg, 0.625µg, 0.3125µg
All lanes: PKM antibody at 1:4000
Secondary
Goat polyclonal to Mouse IgG at 1/10000 dilution
Predicted band size: 58 kDa
Observed band size: 58 KDa
Exposure time: 5min
-
Western Blot
Positive WB detected in: MCF-7 whole cell lysate
All lanes: PKM antibody at 1:4000, 1:8000, 1:16000, 1:32000, 1:64000, 1:128000, 1:256000
Secondary
Goat polyclonal to Mouse IgG at 1/10000 dilution
Predicted band size: 58 kDa
Observed band size: 58 KDa
Exposure time: 5min
-
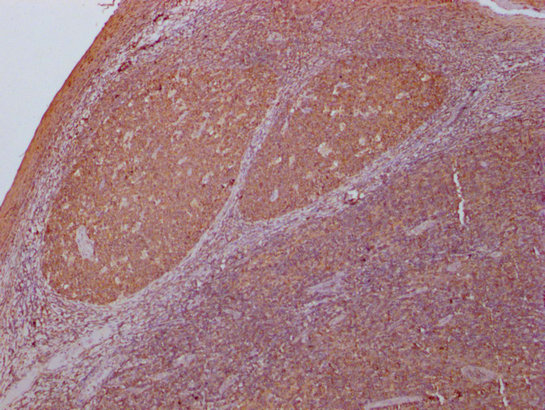
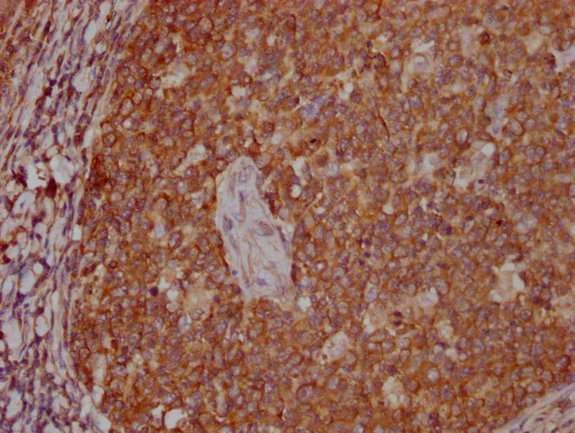
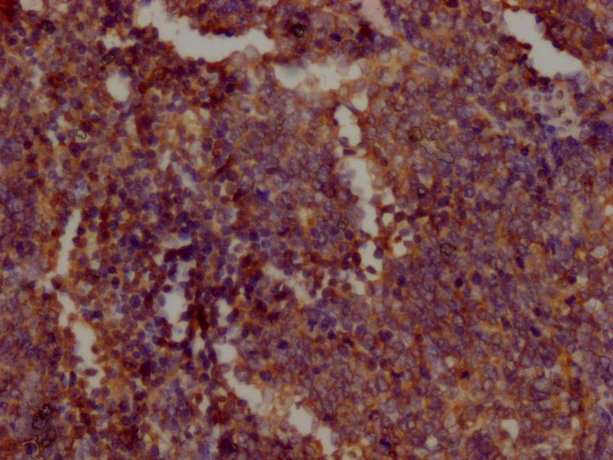
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human tonsil tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human tonsil tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
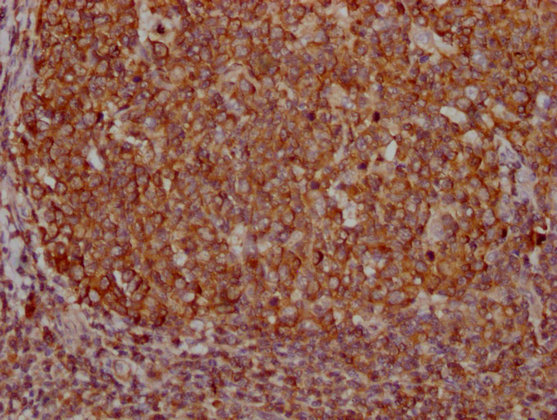
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human tonsil tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human lung cancer tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human lung cancer tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human lung cancer tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
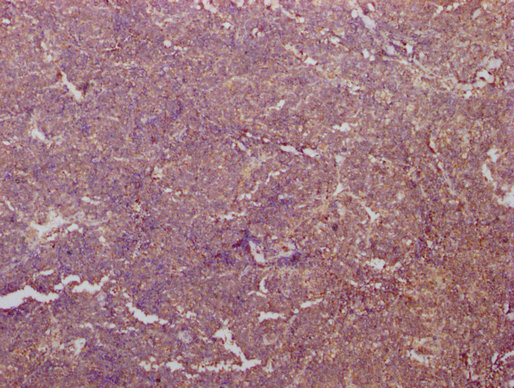
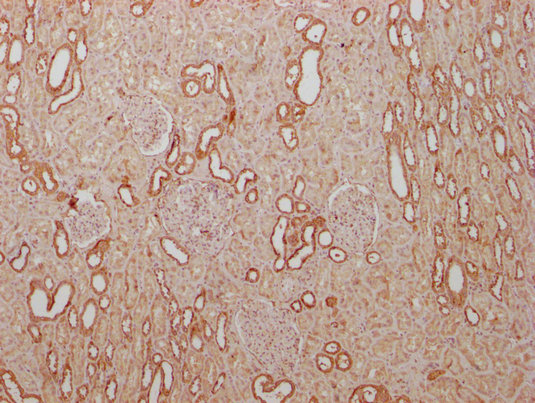
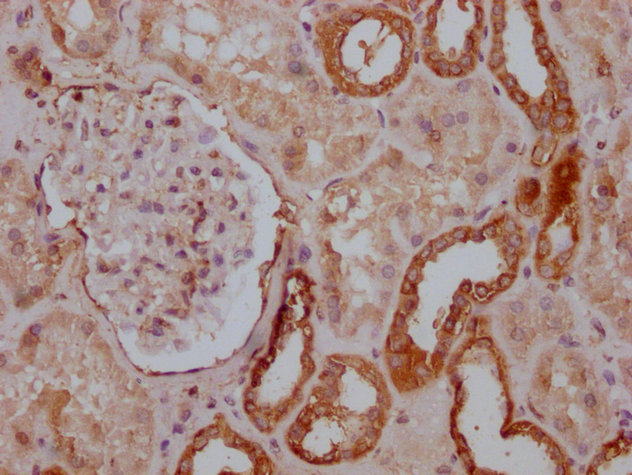
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human kidney tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
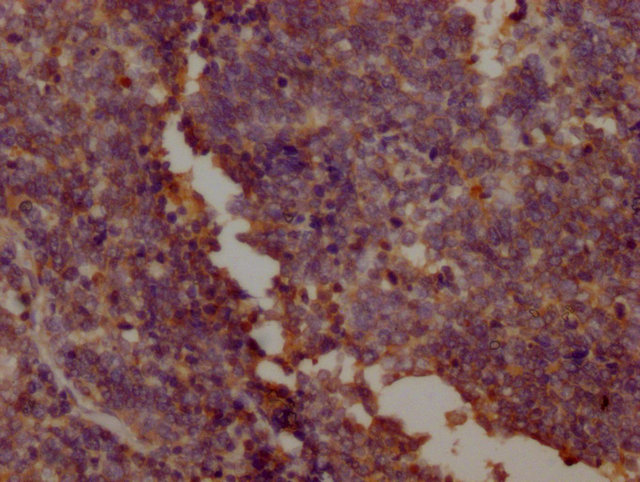
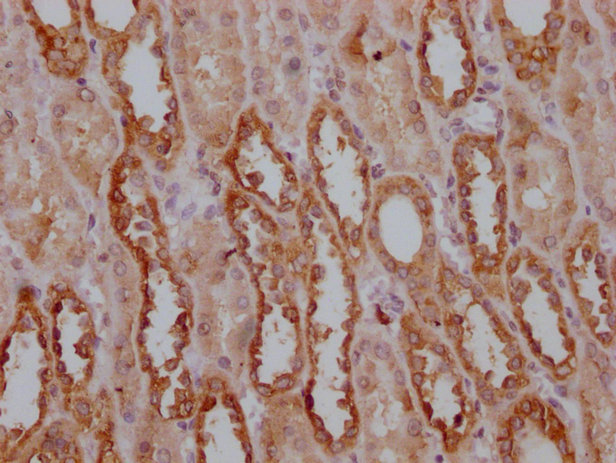
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human kidney tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
IHC image of CSB-MA018072A0m diluted at 1:400 and staining in paraffin-embedded human kidney tissue performed on a Leica BondTM system. After dewaxing and hydration, antigen retrieval was mediated by high pressure in a citrate buffer (pH 6.0). Section was blocked with 10% normal goat serum 30min at RT. Then primary antibody (1% BSA) was incubated at 4°C overnight. The primary is detected by a biotinylated secondary antibody and visualized using an HRP conjugated SP system.
-
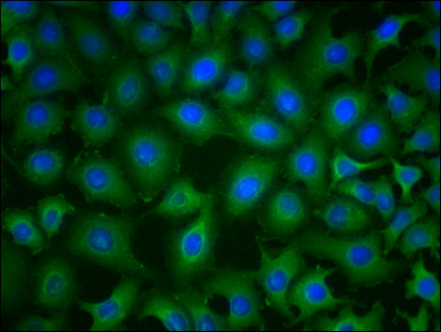
Immunofluorescence staining of A549 cells with CSB-MA018072A0m at 1:230, counter-stained with DAPI. The cells were blocked in 10% normal Goat Serum and then incubated with the primary antibody overnight at 4°C. The secondary antibody was Alexa Fluor 488-congugated AffiniPure Goat Anti-Mouse IgG(H+L).
-
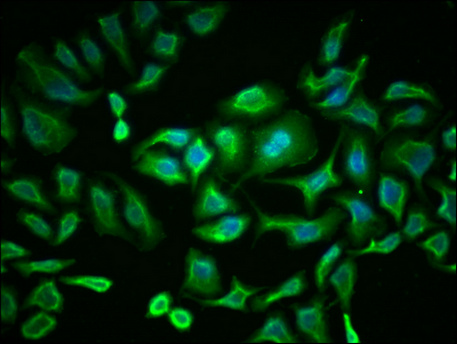
Immunofluorescence staining of Hela cells with CSB-MA018072A0m at 1:230, counter-stained with DAPI. The cells were blocked in 10% normal Goat Serum and then incubated with the primary antibody overnight at 4°C. The secondary antibody was Alexa Fluor 488-congugated AffiniPure Goat Anti-Mouse IgG(H+L).
-
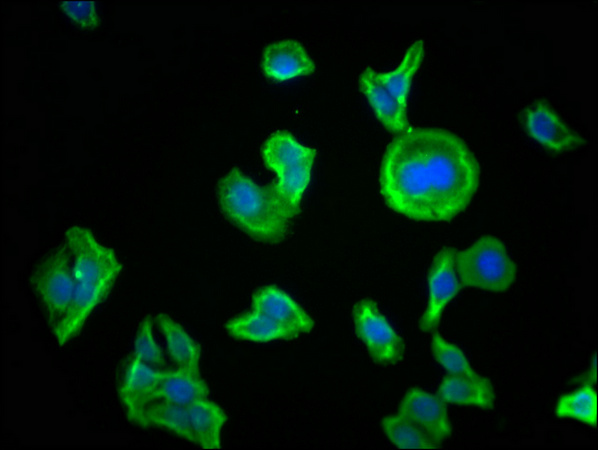
Immunofluorescence staining of HepG2 cells with CSB-MA018072A0m at 1:230, counter-stained with DAPI. The cells were blocked in 10% normal Goat Serum and then incubated with the primary antibody overnight at 4°C. The secondary antibody was Alexa Fluor 488-congugated AffiniPure Goat Anti-Mouse IgG(H+L).
-
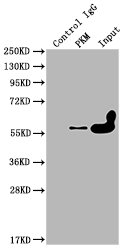
Immunoprecipitating PKM in Hela whole cell lysate
Lane 1: Mouse control IgG instead of CSB-MA018072A0m in Hela whole cell lysate.
Lane 2: CSB-MA018072A0m (1ul) + Hela whole cell lysate (500ug)
Lane 3: Hela whole cell lysate (10ug)
For western blotting, the blot was detected with CSB-MA018072A0m at 1:2000, and a HRP-conjugated Protein G antibody was used as the secondary antibody at 1:2000
-
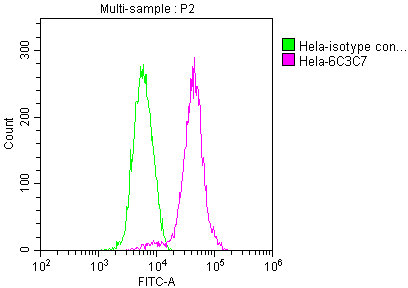
Overlay histogram showing Hela cells stained with CSB-MA018072A0m (red line) at 1:100. The cells were incubated in 1x PBS /10% normal goat serum to block non-specific protein-protein interactions followed by primary antibody for 1 h at 4°C. The secondary antibody used was FITC goat anti-mouse IgG(H+L) at 1/200 dilution for 1 h at 4°C. Isotype control antibody (green line) was used under the same conditions. Acquisition of >10,000 events was performed.
-
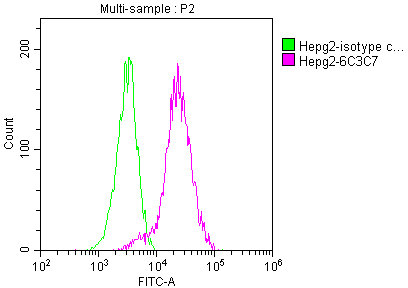
Overlay histogram showing HepG2 cells stained with CSB-MA018072A0m (red line) at 1:100. The cells were incubated in 1x PBS /10% normal goat serum to block non-specific protein-protein interactions followed by primary antibody for 1 h at 4°C. The secondary antibody used was FITC goat anti-mouse IgG(H+L) at 1/200 dilution for 1 h at 4°C. Isotype control antibody (green line) was used under the same conditions. Acquisition of >10,000 events was performed.
|