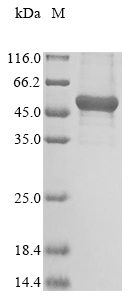
Expressing the recombinant human GTPase NRAS protein in E.coli cells involves inserting a DNA fragment encoding the human NRAS protein (1-186aa) into a plasmid vector along with the N-terminal 10xHis-GST and C-terminal Myc-tag gene and transferring it to the E.coli cells. Positive cells are screened, cultured, and induced to express the NRAS protein. The cells are lysed to collect the recombinant human NRAS protein, which is purified through affinity purification and then identified using SDS-PAGE and subsequent staining of the gel with Coomassie Brilliant Blue. The purity of the resulting recombinant human NRAS protein exceeds 85%.
The NRAS protein, encoded by the neuroblastoma RAS viral oncogene homolog (NRAS) gene, is a crucial player in cellular signaling pathways that regulate processes such as cell proliferation, survival, migration, and differentiation [1]. NRAS is a member of the RAS protein family, which includes KRAS, HRAS, and NRAS, and functions as a molecular switch transmitting signals from activated receptors to the nucleus through intricate downstream signaling cascades [2]. Mutations in NRAS, such as NRAS(Q61), NRAS(G12), and NRAS(G13), can lead to dysregulation of RAS activity, affecting its GTPase function and conformation, thereby contributing to uncontrolled cell growth and proliferation [3].
NRAS is frequently implicated in various cancers, including melanoma, where its mutant forms are challenging to target with conventional therapies [2]. Studies have explored different approaches to modulate NRAS expression and activity. For instance, microRNA-708 has shown promise in reducing NRAS protein levels in cancer cell lines with NRAS mutations, leading to inhibited cell growth and enhanced apoptosis [4]. Additionally, small molecules targeting the NRAS 5' UTR have been investigated as potential inhibitors to counteract the aberrant signaling driven by constitutively active NRAS [5].
Understanding the conformational dynamics of NRAS is crucial for developing targeted therapies. Research has highlighted the importance of the balance of conformational states in influencing the intrinsic hydrolysis of NRAS compared to other RAS isoforms, shedding light on potential vulnerabilities that could be exploited for therapeutic interventions [6]. Moreover, the interaction landscape of RAS paralogs, including NRAS, has been studied to elucidate the proximal protein interactions that drive oncogenic signaling in various cancer types [7].
References:
[1] S. Kumari, A. Bugaut, J. Huppert, & S. Balasubramanian, An rna g-quadruplex in the 5′ utr of the nras proto-oncogene modulates translation, Nature Chemical Biology, vol. 3, no. 4, p. 218-221, 2007. https://doi.org/10.1038/nchembio864
[2] C. Posch and S. Ortiz‐Urda, Nras mutant melanoma - undrugable?, Oncotarget, vol. 4, no. 4, p. 494-495, 2013. https://doi.org/10.18632/oncotarget.970
[3] E. Muñoz‐Couselo, E. Zamora, C. Ortiz, J. García, & J. Peréz-García, <em>nras</em>-mutant melanoma: current challenges and future prospect, Oncotargets and Therapy, vol. Volume 10, p. 3941-3947, 2017. https://doi.org/10.2147/ott.s117121
[4] J. Pang, P. Chien, M. Kao, P. Chiu, P. Chen, Y. Hsuet al., Microrna-708 emerges as a potential candidate to target undruggable nras, Plos One, vol. 18, no. 4, p. e0284744, 2023. https://doi.org/10.1371/journal.pone.0284744
[5] S. Balaratnam, Z. Torrey, D. Calabrese, M. Banco, K. Yazdani, X. Lianget al., Investigating the nras 5’ utr as a target for small molecules,, 2022. https://doi.org/10.1101/2022.01.05.475055
[6] D. Reid and C. Mattos, Balance of conformational states affect the intrinsic hydrolysis of nras when compared to other ras isoforms, The Faseb Journal, vol. 31, no. S1, 2017. https://doi.org/10.1096/fasebj.31.1_supplement.764.14
[7] B. Béganton, E. Coyaud, E. Laurent, A. Mangé, J. Jacquemetton, M. Romanceret al., Proximal protein interaction landscape of ras paralogs, Cancers, vol. 12, no. 11, p. 3326, 2020. https://doi.org/10.3390/cancers12113326