Histone Modifications and DNA Methylation
Epigenetic alterations encompass changes in the DNA structure, stemming from post-replication DNA modifications and post-translational modifications of DNA-associated proteins. Distinguished from mutations, epigenetic changes occur swiftly and are reversible. Among these, DNA methylation and histone protein modifications stand as prominent epigenetic mechanisms, intricately intertwined.
1. Histone Modifications
Histone modifications are post-translational chemical alterations that occur on histone proteins, including acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation, and SUMOylation, which play a crucial role in regulating chromatin structure and gene expression within the cell nucleus. These modifications can either promote or inhibit gene expression by altering the structure of chromatin and are essential for various cellular processes, including transcription regulation, DNA repair, and epigenetic inheritance.
2. DNA Methylation
DNA methylation is an epigenetic modification catalyzed by DNA methyltransferase (DNMT), which transfers an active methyl group from S-adenosyl methionine (SAM) to the fifth position of the pyrimidine ring of cytosine to form 5-methylcytosine (5meC) [5]. DNA methylation changes chromosomal structure, DNA conformation, and DNA stability, thus regulating gene expression either by enlisting proteins responsible for gene suppression or hindering the interaction between transcription factor(s) and DNA.
| Enzymes or proteins Involved in DNA Methylation |
Function |
| Writers |
Dnmt1,
Dnmt3a,
Dnmt3b,
Dnmt3L |
Catalyze the addition of methyl groups onto cytosine residues |
| Erasers |
AID/APOBEC,
TDG,
SMUG1 |
Modify and remove the methyl group |
| Readers |
MBD proteins |
MeCP2,
MBD1,
MBD2, MBD3,
MBD4 |
Recognize and bind to methyl groups to ultimately influence gene expression |
| UHRF proteins |
UHRF1,
UHRF2 |
| Zinc-finger proteins |
Kaiso,
ZBTB4,
ZBTB38 |
In mammals, DNA methylation takes place at cytosines within various genomic contexts [6]. Nevertheless, over 98% of DNA methylation predominantly occurs in cytosine-guanine sequence (CpG) dinucleotide sequences within somatic cells, whereas up to 25% of all methylation events are observed in a non-CpG context within embryonic stem cells (ESCs) [6]. CpG islands, the regions with relatively high concentrations of CpG sequences, are normally in a non-methylated state due to being protected (except for genes located on the inactivated X chromosome and imprinted genes).
DNA methylation is usually erased during zygote formation and subsequently reinstated in the embryo around the time of implantation [7]. Most DNA methylation is crucial for normal development and serves a pivotal role in various vital processes, such as genomic imprinting, X-chromosome inactivation, and the inhibition of transcription and transposition of repetitive elements [5, 8]. Dysregulation of DNA methylation can contribute to diseases like cancer.
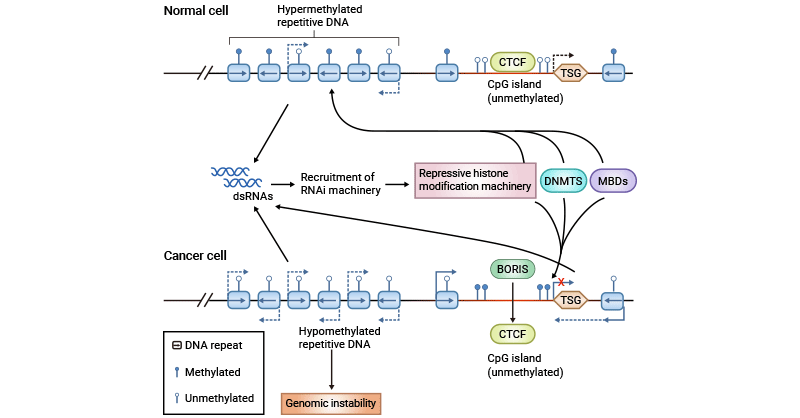
Figure 1. DNA methylation and Cancer
(Note: CTCF, chromatin insulator CCCTC-binding factor; TSG, transcribed tumour suppressor gene; MBDs, methyl-CpG-binding domain-containing proteins; BORIS, brother of regulator of imprinted sites)
The picture is cited from: https://www.nature.com/articles/nrg1655
3. Crosstalk between Histone Modifications and DNA Methylation
In the realm of chromatin, DNA methylation does not operate independently. Instead, an intricate interplay exists between DNA methylation and various histone modifications, encompassing acetylation, methylation, and ubiquitylation.
Preliminary investigations have indicated that the connection between DNA methylation and histone modifications is facilitated through two families of methylated CpG DNA binding proteins: the MBD family (MeCP2 and MBD1-4) and the BTB/POZ family (Kaiso/ZBTB 33 and ZBTB4/38) [1]. These methyl-CpG-binding proteins not only bind to methylated DNA but also associate with multiple distinct chromatin modifying enzymes, including histone deacetylases (HDACs) and histone methyltransferases, thus mediating interactions between DNA methylation and histone modification, generating an inhibitory chromatin structure [2-4].
3.1 Histone Methylation and DNA Methylation
DNA methylation can guide H3K9 methylation through interactions involving DNMTs, H3K9 methyltransferases SUV39H1/2, and methyl-CpG-binding domain proteins [9]. Additionally, the tri-methylation of H3K27 is intricately linked to underlying DNA methylation by H3K27 methyltransferase EZH2-DNMTs direct interaction [10].
Histone modifications can also impact the DNA methylation pattern. Dnmt3L, for instance, associates with H3 histone tails and recruits Dnmt3a and Dnmt3b to initiate DNA methylation [11]. Furthermore, the direct interaction between Dnmt3a and the H3 histone tail, sometimes promoted by the repressive histone mark H3K36m3, boosts its methyltransferase activity [12]. However, the presence of the active histone modification H3K4me3 disrupts the binding of Dnmt3a, Dnmt3b, and Dnmt3L to H3 histone tails, preventing methylation [11].
3.2 Histone Aceylation and DNA Methylation
MBD proteins engage with histone deacetylase (HDAC) enzymes, fostering the creation of transcriptionally repressive chromatin environments by eliminating acetyl groups from lysine residues in histones [13].
Both Dnmt1 and Dnmt3b can interact with HDACs that erase acetylation from histones, resulting in the compaction of DNA, limiting access to transcription [14].
Studies have also shown that enhanced histone acetylation can induce DNA demethylation [15].
3.3 Histone Ubiquitylation and DNA Methylation
Atsuya Nishiyama and colleagues demonstrated that Uhrf1-dependent histone H3 ubiquitylation is a crucial prerequisite for maintaining DNA methylation [16]. Uhrf1 exhibits a particular affinity for hemi-methylated DNA, facilitated by its SRA (SET and RING finger associated) domain, and plays a pivotal role in sustaining DNA methylation by recruiting Dnmt1 to hemi-methylated DNA sites.
Luna Yamaguchi et al. showed that Usp7 is involved in the modulation of maintenance of DNA methylation by deubiquitylating Uhrf1-mediated histone H3 ubiquitination [17]. Jialun Li et al. found that USP7 negatively regulates overall DNA methylation and safeguards the genome from excessive DNA methylation by mitigating the recruitment of histone ubiquitination-dependent DNMT1 [18].
Histone modifications and DNA methylation are intimately linked in epigenetic regulation. These processes collaborate to orchestrate gene expression patterns. Histone modifications, such as methylation and acetylation, shape chromatin structure, influencing DNA methylation by recruiting or repelling DNA methyltransferases. Conversely, DNA methylation can impact histone marks by recruiting proteins that read or erase these modifications.
Together, they establish a dynamic epigenetic landscape that determines gene activity or silencing, playing vital roles in development, disease, and cellular identity. Understanding the interplay between histone modifications and DNA methylation is crucial for unraveling complex gene regulatory networks and their implications in various biological contexts.
References
[1] O. Bogdanovic, G.J. Veenstra. DNA methylation and methyl-CpG binding proteins: developmental requirements and function [J]. Chromosoma, 118 (2009), pp. 549-565.
[2] Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex [J]. Nature. 1998;393:386–389.
[3] Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins [J]. Mol Cell Biol. 1998;18:6538–6547.
[4] Bird A. DNA methylation patterns and epigenetic memory [J]. Genes Dev. 2002;16:6–21.
[5] Robertson KD. DNA methylation and human disease [J]. Nat Rev Genet. 2005;6(8):597-610.
[6] Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences [J]. Nature. 2009; 462(7271):315-22v.
[7] Zhu JK. Active DNA demethylation mediated by DNA glycosylases [J]. Annu Rev Genet. 2009;43:143-66.
[8] Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease [J]. Mutat Res. 2008;647(1-2):30-8.
[9] Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin [J]. Curr Biol. 2003;13(14):1192-200.
[10] Vire E, Brenner C, Deplus R, et al. The polycomb group protein EZH2 directly controls DNA methylation [J]. Nature. 2006;439(7078):871-4.
[11] Ooi SK, Qiu C, et al. (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA [J]. Nature 448: 714–717.
[12] Dhayalan A, Rajavelu A, et al. (2010) The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation [J]. J Biol Chem 285: 26114–26120.
[13] Ng HH, Zhang Y, et al. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex [J]. Nat Genet 23: 58–61.
[14] Geiman TM, Sankpal UT, et al. (2004) DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system [J]. Biochem Biophys Res Commun 318: 544–555.
[15] Cervoni N, Szyf M (2001). Demethylase activity is directed by histone acetylation [J]. J Biol Chem 276: 40778–40787.
[16] Nishiyama, A. et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication [J]. Nature 502, 249–253 (2013).
[17] Yamaguchi, L., Nishiyama, A., Misaki, T. et al. Usp7-dependent histone H3 deubiquitylation regulates maintenance of DNA methylation [J]. Sci Rep 7, 55 (2017).
[18] Li, J., Wang, R., Jin, J. et al. USP7 negatively controls global DNA methylation by attenuating ubiquitinated histone-dependent DNMT1 recruitment [J]. Cell Discov 6, 58 (2020).