Histone ADP-ribosylation
It wasn't until the late 1990s and early 2000s that researchers began to discover histone ADP-ribosylation as a specific modification. Studies showed that PARP enzymes could ADP-ribosylate histone proteins, particularly histone H1 and histone H2B. Subsequent research has demonstrated extensive roles of histone ADP-ribosylation.
1. What Is Histone ADP-ribosylation?
The process in which the ADP-ribose (ADPr) group from the co-factor nicotine-amide adenine dinucleotide (NAD+) is transferred to specific amino-acids (Lys, Arg, Glu, Asp, and Ser) on histone proteins is known as histone ADP-ribosylation.
ADP-ribosylation emerges as one of the earliest damage signals produced at DNA lesions. Histones serve as the major substrates of ADP-ribosylation in response to DNA damage because they are proximal to DNA [1,2].
All five histone proteins are extensively ADP-ribosylated, albeit in a small percentage. Histone ADP-ribosylation is proposed as a covalent alteration [8], primarily involving ADP-ribose monomers or chains or branched polymers [9]. The ADP-ribosylation pattern of histone proteins differs based on the chromatin composition. In native chromatin, histone H1 serves as the primary ADP-ribose acceptor such as H1E2 and H1E15, whereas in H1-depleted chromatin, histone H2B becomes the most heavily ribosylated histone [10].
2. Components Involved in Histone ADP-ribosylation
Histone ADP-ribosylation is a dynamic and reversible enzymatic process regulated by three key categories of proteins: writers, erasers, and readers.
2.1 Writers
Enzymes that catalyze ADP-ribosylation modification of histones are ADP-ribosyltransferases (ARTs) or poly(ADP-ribose) polymerases (PARPs), also called ADP-ribose "writers", which possess mono- or poly-ADP-ribose transferase activity, respectively.
There are three families of ADP-ribosyltransferases, including the diphtheria toxinlike ADP-ribosyltransferases (ARTDs, formerly known as PARPs) with both mono- and poly-ADP-ribosyltransferase activities, the clostridial toxinlike ADP-ribosyltransferases (ARTCs) and the Sir2 family of NAD+-dependent protein deacetylases (sirtuins). The latter two exclusively function as mono-ADP-ribosyltransferases.
| ADP-ribosyltransferases |
Sub-type |
Subcellular location |
Enzymatic activities |
| ARTDs |
ARTD1 |
Nucleus |
Poly-ADP-ribosyltransferase |
| ARTD2 |
Nucleus |
Poly-ADP-ribosyltransferase |
| ARTD3 |
Nucleus |
Mono/Poly-ADP-ribosyltransferase |
| ARTD4 |
Nucleus/cytoplasm |
Poly-ADP-ribosyltransferase |
| ARTD5 |
Nucleus/cytoplasm |
Poly-ADP-ribosyltransferase |
| ARTD6 |
Nucleus/cytoplasm |
Poly-ADP-ribosyltransferase |
| ARTD7 |
Nucleus |
Mono/Poly-ADP-ribosyltransferase |
| ARTD8 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase |
| ARTD9 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase |
| ARTD10 |
Nucleus/cytoplasm |
Mono/Poly-ADP-ribosyltransferase |
| ARTD11 |
- |
Mono/Poly-ADP-ribosyltransferase |
| ARTD12 |
Nucleus |
Mono/Poly-ADP-ribosyltransferase |
| ARTD13 |
Nucleus |
Catalytically inactive |
| ARTD14 |
Nucleus |
Mono-ADP-ribosyltransferase |
| ARTD15 |
Nucleus/cytoplasm |
Mono-ADP-ribosyltransferase |
| ARTD16 |
- |
Mono-ADP-ribosyltransferase |
| ARTD17 |
- |
Mono-ADP-ribosyltransferase |
| ARTCs |
ARTC1 |
Ecto-cellular |
- |
| ARTC2 |
Ecto-cellular |
- |
| ARTC3 |
Ecto-cellular |
- |
| ARTC4 |
Ecto-cellular |
- |
| ARTC5 |
Ecto-cellular |
- |
| Special ARTs |
ADPRT1a |
- |
Mono-ADP-ribosyltransferase |
| ADPRT2 |
- |
Mono-ADP-ribosyltransferase |
| Sirtuins |
SIRT1 |
Nucleus |
- |
| SIRT2 |
Nucleus/Cytoplasm |
- |
| SIRT3 |
Mitochondria |
- |
| SIRT4 |
Mitochondria |
Mono-ADP-ribosyltransferase |
| SIRT5 |
Mitochondria |
- |
| SIRT6 |
Nucleus |
Mono-ADP-ribosyltransferase |
| SIRT7 |
Nucleus (nucleoli) |
Mono-ADP-ribosyltransferase |
The table information is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8027728/
| Substrate |
Modified amino acid |
Enzymes |
Effect of the reaction |
| H1 |
E2, E14, E116 and K213 |
- |
Regulation of H1-H1 interactions |
| R34 |
- |
Blocks the cAMP-dependent phosphorylation of histone H1 |
| Q/N |
ARTD3 |
DNA repair |
| H2A |
Unknown |
Sir2 |
Response to oxidative stress/DNA damage |
| |
Unknown |
Sir2 |
Inhibition of histone acetylation/silencing chromatin domains |
| |
R/E |
- |
Unknown |
| |
K13 |
ARTD1 |
Unknown |
| H2B |
E2 |
ARTD3/ARTD10 |
Unknown |
| |
E18/E19 |
ARTs, Adprt1a/Adprt2 |
Response to oxidative stress/DNA damage |
| |
Unknown |
Sir2 |
Inhibition of histone acetylation/silencing of chromatin domains |
| |
K30 |
ARTD1 |
Unknown |
| H3 |
Unknown |
SIRT6, Sir2 |
Inhibition histone acetylation/silencing chromatin domains |
| |
R |
- |
Cell proliferation |
| |
K27 |
ARTD1 |
Unknown |
| |
K37 |
ARTD1 |
Unknown |
| H4 |
R |
Sir2 |
Post-synthetic modification with acetylation of core histones |
| |
Unknown |
Sir2 |
Response to oxidative stress/DNA damage |
| |
Unknown |
ARTD10 |
Unknown |
| |
Unknown |
Sir2-relatedprotein |
Inhibition histone acetylation |
| |
K16 |
ARTD1 |
Unknown |
The information is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8027728/
(Note: E, glutamate; K, lysine; R, arginine; Q, glutamine; N, asparagine)
2.2 Erasers
Terminal ADP-ribosyl glycohydrolases (TARG) and poly-ADP-ribose glucohydrolases (PARG), also known as "erasers", are responsible for removing ADP-ribose monomer or polymers from ADP-ribosylated histones, respectively.
2.3 Readers
The ADP-ribose modification is recognized by proteins that contain poly-ADP-ribose binding zinc finger (PBZ) domains or macrodomains, which subsequently modulate chromatin structure and transcription.
| Reader Module |
|
Function |
| PBM |
Histone H2A, Histone H2B, Histone H3, Histone H4 |
DNA repair, chromatin rearrangements, transcription, gene expression |
| Macrodomain |
macroH2A1.1 |
Chromatin remodeling, DNA repair |
3. The Mechanism of Histone ADP-Ribosylation
Histone ADP-ribosylation involves a complex series of molecular events. Here's a simplified overview of the process:
3.1 Recognition of DNA Damage
One of the best-studied roles of histone ADP-ribosylation is in DNA repair. When the cell detects DNA damage, such as breaks or lesions, PARP enzymes are activated and then bind to the damaged DNA and begin synthesizing ADP-ribose chains on nearby histones.
3.2 Chromatin Relaxation
The addition of ADP-ribose to histones results in chromatin relaxation, making the damaged DNA more accessible to repair machinery. This step is essential for efficient DNA repair.
3.3 Recruitment of Repair Proteins
The ADP-ribose chains act as a signal for DNA repair proteins to assemble at the site of damage. These proteins then carry out the necessary repairs, ensuring the integrity of the genome.
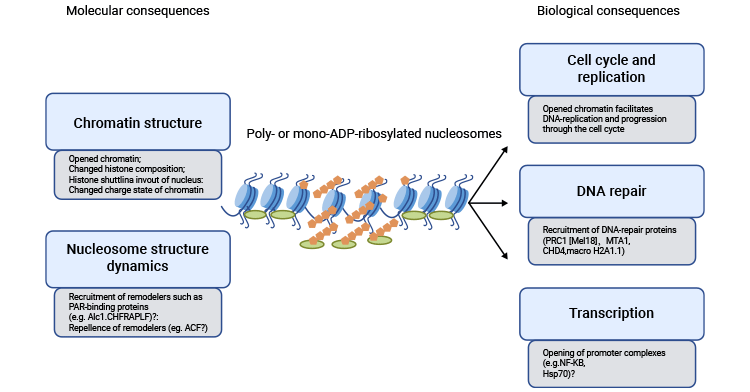
4. Function of Histone ADP-ribosylation
Numerous studies have demonstrated the significant contributions of histone poly-ADP-ribosylation in DNA repair and DNA replication [2, 3], as well as in the proliferation of both cells and tumors [4]. Histone mono-ADP-ribosylation also contributes to transcription and chromatin remodeling [7].
Figure 1. Histone ADP-ribosylation Function
This picture is cited from: https://www.sciencedirect.com/science/article/abs/pii/S0962892411001061
5. Crosstalk between Histone ADP-ribosylation and Other Histone Modifications
Interactions among various PTMs can occur either by directly competing for acceptor sites or indirectly by altering the chromatin accessibility for modifying enzymes. The finding that specific lysine residues can act as ADP-ribose acceptors is noteworthy since these same amino acid residues are potential targets for acetylation and methylation [5]. Consequently, it is probable that competition for receptor sites among ADP-ribosylation, acetylation, methylation, and phosphorylation, leads to crosstalk [3]. This notion is substantiated by findings showing that H4 K16ac represses ADP-ribosylation in vitro [6], implying the existence of similar crosstalk in vivo.
References
[1] Ogata N, Ueda K, and Hayaishi O (1980) ADP-ribosylation of histone H2B-identification of glutamic acid residue 2 as the modification site [J]. J. Biol. Chem 255, 7610–7615.
[2] Messner S, Hottiger MO (2011) Histone ADP-ribosylation in DNA repair, replication and transcription [J]. Trends Cell Biol 21: 534–542.
[3] Hottiger MO. ADP-ribosylation of histones by ARTD1: an additional module of the histone code [J]? FEBS Lett. 2011; 585:1595–99.
[4] Boulikas T, Bastin B, Boulikas P, Dupuis G. Increase in histone poly (ADP-ribosylation) in mitogen-activated lymphoid cells [J]. Exp Cell Res. 1990; 187:77–84.
[5] Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705.
[6] Messner S, Altmeyer M, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362.
[7] Boulikas T. Relation between carcinogenesis, chromatin structure and poly(ADP-ribosylation) (review) [J]. Anticancer Res. 1991;11:489–527.
[8] P. Stone, W. Lorimer, W. Kidwell. Properties of the complex between histone H1 and poly(ADP-ribose synthesised in HeLa cell nuclei [J]. Eur. J. Biochem., 81 (1977), pp. 9-18.
[9] L. Burzio, P. Riquelme, S. Koide. ADP ribosylation of rat liver nucleosomal core histones [J]. J. Biol. Chem., 254 (1979), pp. 3029-3037.
[10] A. Huletsky, G. de Murcia, et al. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure [J]. J. Biol. Chem., 264 (1989), pp. 8878-8886.