Histone Variants
In 1977, Franklin and Zweidler initially isolated histone variants from mammalian tissues using acid-urea-Triton X (AUT) polyacrylamide gel electrophoresis [1]. Histones exhibit a wide array of functions encompassing gene expression, chromosome segregation, DNA repair, and other fundamental chromosomal processes in eukaryotes. The unique demands of these chromosomal processes have driven the emergence of specific histone variants. The integration of such variant histones into nucleosomes gives rise to tremendous diversity in chromatin structure.
1. What Are Histone Variants?
Histone variants are nonallelic isoforms of core histones and replace their canonical histone counterparts at strategic locations in the genome. Histone variants differ from core histones varying from minor alterations in amino acids to substantial structural differences. Canonical histones are typically synthesized during genome replication with peak expression in S-phase, whereas histone variants are expressed across the cell cycle and incorporated into chromatin in a replication-independent manner. Some histone variants exhibit tissue-specific expression, with restriction to germ cells.
Typically, eukaryotic genomes house replication-coupled (RC) histones in large multi-copy arrays, while histone variants are found in limited quantities, often as a single or just a few copies. Although histone variants share similarities with their RC histone counterparts, they exhibit distinct characteristics in their histone-fold domains (HFDs), setting them apart in terms of sequence and function.
Additionally, histone variants frequently display more significant differences from RC histones in their N- and C-terminal tails. These distinctions result in their association with different chaperones for deposition and the imposition of distinct post-translational modifications. As a consequence, these variations endow histone variants with specialized functions through alteration in chromatin properties [4,5].
2. Classifications of Histone Variants
All histones can exist as variants, but the diversification of replication-coupled (RC) histones into histone variants is not uniform. In the case of mammals, there are many H2A variants, a relatively smaller number of H3 variants, and even fewer H2B and H4 variants. This uneven diversification could be linked to the position of each histone within the nucleosome and its potential to modify nucleosome properties upon substitution.
| Canonical histones |
Histone variants |
Function |
| H2A |
H2A.X |
Distributed throughout the genome, DNA repair, remodeling sex chromosomes |
| H2A.Z |
H2A.Z.1 |
Transcriptional regulation, heterochromatin organization, chromosome segregation, genome stability, cellular proliferation |
| H2A.Z.2 |
Transcriptional regulation |
| H2A.Bbd |
Restricted to vertebrates, excluded from the inactive X, transcriptional activation |
| macroH2A1.2 |
Restricted to vertebrates, located in inactive X chromosome in female mammalian cells, transcription suppression and higher-order chromatin compaction |
| H2A.J |
Gene expression related to cell proliferation and inflammation |
| H2A.B |
Nucleosome destablization, active transcription, DNA repair, DNA replication, splicing, spermatogenesis |
| mouse H2A.L (H2A.L.2) |
Spermatogenesis, histone-to-protamine transition shown for H2A.L2 |
| H2A.P |
Testis-specific |
| H2A.W |
plant-specific; protect an additional 10-15 bp of linker DNA beyond the 147 bp of most nucleosomes from micrococcal nuclease |
| H2B |
H2B.E |
Odour-sensing neurons of mice |
| H2B.W |
Testis-specific |
| TH2B |
Testis-specific, histone-to-protamine transition |
| H2B.3 |
Enriched in mature leaves and in nucleosomes containing H3.3 and/or H2A.Z |
| H2B.8 |
Enriched in dry seed |
| H3 |
H3.1 |
Canonical, replication dependent |
| H3.2 |
Canonical, replication dependent |
| H3.3 |
Cell cycle independent, transcriptional activation, telomere, centromere |
| H3T (H3.4)
|
Testis-specific, expressed in spermatocytes and spermatids, involved in the dynamics of chromatin transtions during spermatogenesis |
| mouse H3t |
Spermatogenesis |
| H3.5 |
localized in the 12p11.21 region of human chromosome 12, mainly expressed in spermatogonia and primary spermatocytes of germinal stages VI-X, spermatogenesis, transcription regulation, |
| H3.6 |
Weakly expressed in various tissues |
| H3.7 |
Weakly expressed in various tissues |
| H3.8 |
Weakly expressed in various tissues |
| H3.X |
located at the 5p15.1 chromosome region in humans, transcriptional regulation, regulate cell cycle |
| H3.Y |
located at the 5p15.1 chromosome region in humans, transcriptional regulation, regulate cell cycle |
| CENP-A (CenH3) |
Centromere identity, required for kinetochore assembly and chromosome segregation during mitosis |
| H3mm7 |
Mouse H3.3 sub-variant, expressed in skeletal muscle satellite cells |
| H4 |
H4-G |
Upregulation of rDNA transcription |
| H1 |
H1.1 |
- |
| H1.2 |
In mouse embryonic stem cells, H1.2 and H1.3 are enriched in H3K9me3-marked heterochromatic domains |
| H1.3 |
| H1.4 |
|
| H1.5 |
In human lung fibroblasts, H1.5 is enriched over splice sites of exons shorter than nucleosomes, promoting their inclusion by stalling RNAPII |
| H1.6 |
Testis-specific |
| H1T2 |
- |
| H1oo |
- |
| HILS1 |
- |
| H1X |
Enriched at RNAPII-enriched domains; an adverse prognosticator for astrocytic gliomas |
| H1.0 |
Enriched at nucleolus-associated domains; often heterogeneously expressed in tumor cells, with H1.0 levels correlating with tumor cell differentiation and patient survival |
Table 1. Mammalian histone variants.
The Table information is cited from: https://www.sciencedirect.com/science/article/pii/S0022283620305866 and https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8015243/
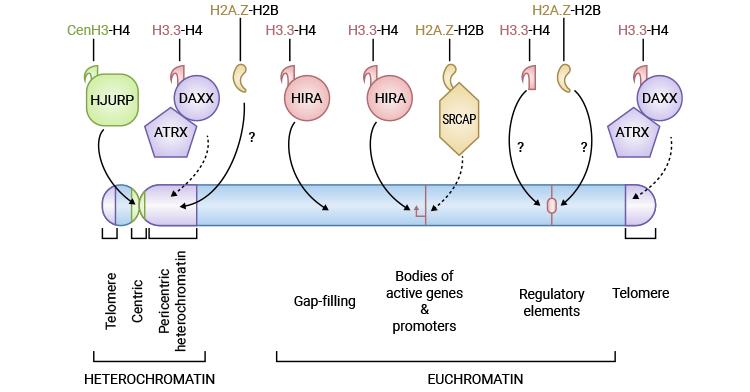
Figure 1. Properties of Histone Variants
3. Function of Histone Variants
Histone variants affect nucleosome stability and higher-order chromatin organization by impacting histone post-translational modifications and the recruitment of histone variant-specific binding partners [3,6]. Histone variants play pivotal roles in nearly all DNA-templated processes, such as DNA repair, chromosome segregation, transcription regulation, genome stability, epigenetic inheritance, and even tissue-specific functions [2,3].
For instance, the centromere-specific variant centromeric protein A (CENP-A) serves as an epigenetic marker for the centromeric region of chromosomes, while H3.3 is closely linked to chromatin dynamics and nucleosome turnover. Moreover, numerous distinct H2A variants are involved in DNA repair, gene regulation, and other critical processes, thus affecting DNA organization. Therefore, the dysregulation of histone variant expression and deposition would lead to developmental syndromes and cancer.
4. Histone Variants and Diseases
Since histone variants have universal expression and pleiotropic effects, it is not surprising that they are implicated in normal physiology and many diseases, including developmental diseases and cancer. For instance, since histone variants may be the sole source of new histones in post-mitotic terminally differentiated tissues, such as the brain, histone variants may contribute significantly to brain-associated diseases, such as neurodevelopmental and neurodegenerative diseases. In fact, some studies suggest that the H3.3 and H2A.Z variants are essential for brain function [7-9].
In addition, given that many histone variants are testis-specific, replacement or modification of testis-specific histone variants might lead to male infertility [10].
It is known that many cancers and developmental syndromes carry recurrent mutations in genes encoding chromatin-related proteins, including histone variants and their deposition-specific partners. Studies have shown that mutations in H3.3 are frequently observed in pediatric gliomas and chondroblastomas [11, 12]. H2A.Z are demonstrated to exert oncogenic functions, while macroH2A and H2A.X inhibit tumorigenesis. Upregulation of H2A.Z is found in prostate and bladder cancer [13, 14]. The expression of MacroH2A declines as melanoma, bladder cancer, and anal neoplasms advance in disease progression [15, 16].
References
[1] S.G. Franklin, A. Zweidler. Non-allelic variants of histones 2a, 2b and 3 in mammals [J]. Nature, 266 (1977), pp. 273-275.
[2] Talbert PB, Henikoff S. 2010. Histone variants--ancient wrap artists of the epigenome [J]. Nat Rev Mol Cell Biol 11:264–275.
[3] Martire S, Banaszynski LA. 2020. The roles of histone variants in fine-tuning chromatin organization and function [J]. Nat Rev Mol Cell Biol 21:522–541.
[4] Henikoff S, Smith MM. 2015. Histone variants and epigenetics [J]. Cold Spring Harb Perspect Biol 7:a019364.
[5] Talbert PB, Henikoff S. 2017. Histone variants on the move: substrates for chromatin dynamics [J]. Nat Rev Mol Cell Biol 18:115–126.
[6] Buschbeck M., Hake S.B. Variants of core histones and their roles in cell fate decisions, development and cancer [J]. Nat. Rev. Mol. Cell Biol. 2017;18:299–314.
[7] Xia W & Jiao J Histone variant H3.3 orchestrates neural stem cell differentiation in the developing brain [J]. Cell Death Differ. 24, 1548–1563 (2017).
[8] Stefanelli G et al. Learning and age-related changes in genome-wide H2A.Z binding in the mouse hippocampus [J]. Cell Rep. 22, 1124–1131 (2018).
[9] Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM & David Sweatt J Histone H2A.Z subunit exchange controls consolidation of recent and remote memory [J]. Nature 515, 582–586 (2014).
[10] Govin J et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis [J]. J. Cell Biol 176, 283–294 (2007).
[11] Schwartzentruber J et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma [J]. Nature 482, 226–231 (2012).
[12] Behjati S et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone [J]. Nat. Genet 45, 1479–1482 (2013).
[13] Dryhurst D., Ausió J. Histone H2A.Z deregulation in prostate cancer. Cause or effect [J]? Cancer Metastastis Rev. 2014;33:429–439.
[14] Kim K., Punj V., et al. Gene dysregulation by histone variant H2A.Z in bladder cancer [J]. Epigenetics Chromatin. 2013;6:1–13.
[15] Kapoor A., Goldberg M.S., et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8 [J]. Nature. 2010;468:1105–1109.
[16] Hu W.-H., Miyai K., et al. Loss of histone variant macroH2A2 expression associates with progression of anal neoplasm [J]. J. Clin. Pathol. 2016;69:627–631.