Histone SUMOylation
In 2003, histone SUMOylation was first described in human HEK293T and P493-6 B cells by Shiio and Eisenman and was shown to cause transcriptional repression [1]. Subsequently, histone SUMOylation was also reported in yeast [2], parasitic protozoans [3], and plants [4].
1. What Is Histone SUMOylation?
Histone SUMOylation refers to the process in which a small ubiquitin-like modifier (SUMO) moiety is covalently linked to a lysine (K) residue on the histone protein.
Like histone ubiquitylation, all core histones, the linker histone H1, the histone variants H2A.Z and H2A.X and the centromeric histone variant Cse4 can be SUMOylated in yeast [5-7]. In mammals, SUMOylation mainly occurs on H4, and H4 SUMOylation induces transcriptional repression [1,8]. Histone SUMOylation currently stands as the sole described repressive histone mark in budding yeast owing to the lack of repressive lysine methylation marks like H3K9me and H3K27me.
2. Three Components Involved in Histone SUMOylation
Histone SUMOylation is a reversible process regulated by several enzymes and proteins that act as writers, readers, and erasers.
2.1 Writers
Histone SUMOylation writers are enzymes responsible for adding the SUMO motif to histones.
2.2 Readers
Histone SUMOylation readers are proteins that recognize and bind to SUMOylated histones, thus translating the signal into different biological outputs. Many proteins have specific SUMO-interacting motifs (SIMs) that enable them to bind to SUMO-modified proteins, including SUMOylated histones. These SIM-containing proteins can include transcription factors, chromatin remodelers, and other regulatory proteins.
| Readers |
SUMOylated histones |
| HP1/CBX Proteins (HP1α/CBX5,
HP1β/CBX1,
and HP1γ/CBX3) |
Bind to SUMOylated histones, particularly at heterochromatin regions; involved in heterochromatin maintenance |
| TIF1 (TIF1α/TRIM24 and
TIF1β/TRIM28) |
Interact with SUMO-modified histones and play a role in transcriptional regulation |
| PML |
Interacts with SUMO-modified histones, contributing to the regulation of gene expression and genome stability |
| SETDB1 |
Interact with SUMOylated histones in the context of heterochromatin formation |
| ATRX |
Interacts with SUMO-modified histones and is implicated in the regulation of heterochromati |
| BRCA1 |
Recognize SUMOylated histones at sites of DNA damage |
| PCNA |
Interacts with SUMO-modified histones during DNA replication and repair processes |
| BRD4 |
Interact with SUMO-modified histones in the context of transcriptional regulatio |
2.3 Erasers
Histone sumoylation can be reversed by deSUMOylaes, also called erasers, including the specific cysteine-protease, Sentrin-specific protease (SENP), which removes the sumoylation marks on histone proteins.
| DeSUMOylases |
Removal of SUMO |
| SENP-1 |
SUMO-1, SUMO-2/3 |
| SENP-2 |
SUMO-1, SUMO-2/3 |
| SENP-3 |
SUMO-2/3 |
| SENP-5 |
SUMO-2/3 |
| SENP-6 |
polySUMO-2/3 (SUMO chain) |
| SENP-7 |
polySUMO-2/3 (SUMO chain) |
| USPL1 |
SUMO-2/3 |
| DeSI1 |
SUMO-2/3 |
| DeSI2 |
SUMO-2/3 |
3. Mechanism of Histone SUMOylation
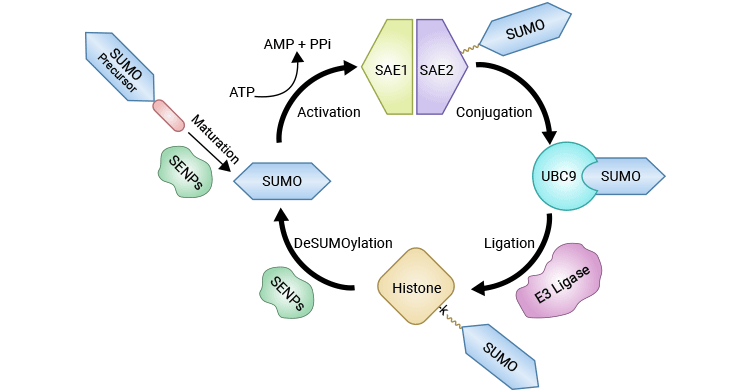
Histone SUMOylation is coordinated through a multi-step enzyme-dependent reaction, involving SUMO E1 activating enzymes, E2 conjugating enzymes, and E3 ligases. All SUMO proteins including SUMO1-4, are firstly catalyzed into mature forms, the C-terminal diglycine of which is essential for E1-mediated adenylation. The E1 enzyme, a heterodimer of SAE1 and SAE2 subunits, triggers the activation of SUMO proteins in an ATP-dependent manner, forming the E1-SUMO intermediate. Once activated, SUMO is subsequently transferred from the E1-SUMO intermediate to the E2 conjugating enzyme UBC9 through a transesterification reaction. Finally, the E3 ligases PIAS (Protein Inhibitor of Activated STAT) and RanBP2 (Ran Binding Protein 2) facilitate to transfer of the SUMO from UBC9 to the specific lysine residue on the targeted histone.
Figure 1. The catalytic cycle of histone SUMOylation and deSUMOylation
The picture is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9654019/
4. Function of Histone SUMOylation
Histone SUMOylation has diverse functions in cotranscriptional processes, including transcription regulation, chromatin remodeling, transcript elongation, and blocking cryptic initiation.
Histone SUMOylation can cause changes in nucleosome structure, which leads to chromatin remodeling and affects the binding of various transcription factors to DNA, thus inhibiting the transcription of certain genes.
Several studies have shown that histone SUMOylation leads to a reduction of transcriptional activity. For example, histone 4 sumoylation mediates transcriptional repression through the recruitment of HDACs and heterochromatin protein 1 (HP1). Interestingly, it has been recently shown that sumoylation regulates the enzymatic activity of histone-modifying enzymes, such as HDAC1, HDAC2, HDAC4, SIRT1, EZH2, and KDM5B, thereby regulating also indirectly the chromatin state.
Additionally, SUMO2/3 modification of the linker histone H1 promoted its binding to highly condensed heterochromatin in ESCs. In contrast, the absence of sumoylation caused chromatin decompaction and the restoration of totipotency [9].
Table 1. Histone SUMOylation sites and functions
| Organism |
Histone |
SUMOylated Sites |
Function |
| Human |
H2A |
- |
Transcriptional repression or chromatin compaction |
| H3 |
K18 |
- |
| H4 |
K12 |
- |
| H2AX |
K5, K9, K13, K15, K118, K119, K127, K133, K134 |
- |
| S. cerevisiae |
H2A |
K126 |
Transcriptional repression/activation, inhibition of cryptic initiation |
| H2B |
K6, K7, K16, K17 |
- |
| H3 |
|
- |
| H4 |
K5, K8, K12, K16, K20 |
- |
| H2A.Z |
K126, K133 |
DSB repair |
| Cse4 |
K65, K215, K216 |
Cse4 incorporation or proteolysis |
The information is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8216275/
5. Crosstalk between Histone SUMOylation and Other Histone Modifications
SUMOylation of H4 increases its interaction with histone deacetylase HDAC1 and the heterochromatin protein 1 HP1γ, which is in agreement with the role of SUMO in transcriptional repression [1]. SUMO has been shown to be necessary for the deposition of the H3K9me3 molecular hallmark of heterochromatin associated with gene silencing [10,11].
Nathan and his team have elegantly employed a blend of chemical, biochemical, and genetic methods to present compelling proof that histone SUMOylation opposes their acetylation, establishing itself as an evolutionarily conserved repressive mark [4]. In S. cerevisiae, histone SUMOylation is shown to antagonize certain activating modifications, such as acetylation and H2B ubiquitylation (maybe on the same lysine residues) [4]. The Chatterjee lab has found that H4K12su stimulates demethylase and deacetylase activities within the transcriptionally repressive LSD1-CoREST1-HDAC1 complex.
Many SUMOylated lysines can be subjected to methylation, acetylation, or ubiquitination. Crosstalk between SUMOylation and phosphorylation occurs during the cell cycle for a regulated cell cycle progression.
6. Histone Sumoylation and Diseases
Accumulating evidence suggests that histone SUMOylation plays a critical role in embryonic development, cellular differentiation, and disease states. Dysregulation of SUMOylation processes has been associated with various cancers, neurodegenerative disorders, and immunological diseases. Histone SUMOylation is very important for the maintenance of histone function and DNA transcription, and its imbalance will affect the cell cycle, differentiation, and apoptosis, and may lead to the development of tumors.
References
[1] Shiio Y, Eisenman RN. 2003 Histone sumoylation is associated with transcriptional repression [J]. Proc. Natl Acad. Sci. USA 100, 13 225-13 230.
[2] Ryu, H. Y., Su, D., et al. (2019) The Ulp2 SUMO protease promotes transcription elongation through regulation of histone sumoylation [J]. EMBO J. 38, e102003.
[3] Issar, N., Roux, E., Mattei, D., and Scherf, A. (2008) Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments [J]. Cell. Microbiol. 10, 1999–2011.
[4] Miller, M. J., Barrett-Wilt, et a. (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis [J]. Proc. Natl. Acad. Sci. U.S.A. 107, 16512–16517.
[5] Nathan, D., Ingvarsdottir, K., et a. (2006) Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications [J]. Genes Dev. 20, 966– 976.
[6] Kalocsay M, Hiller NJ, Jentsch S (2009) Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break [J]. Mol Cell 33: 335–343.
[7] Ohkuni K., Takahashi Y., F, et a. SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin [J]. Mol. Biol. Cell. 2016; 27:1500–1510.
[8] Shiio, Y., and Eisenman, R.N. (2003). Histone sumoylation is associated with transcriptional repression [J]. Proc Natl Acad Sci U S A 100, 13225–30.
[9] Sheban, D., Shani, T., et a. (2022). SUMOylation of linker histone H1 drives chromatin condensation and restriction of embryonic cell fate identity [J]. Mol. Cell. 82, 106–122.e9.
[10] Ninova M, Godneeva B, et al. 2020 The SUMO Ligase Su(var)2–10 controls hetero- and euchromatic gene expression via establishing H3K9 trimethylation and negative feedback regulation [J]. Mol. Cell 77, 571-585.e4.
[11] Ninova M, Fejes Toth K, Aravin AA. 2019 The control of gene expression and cell identity by H3K9 trimethylation [J]. Development 146, dev181180.