Full Product Name
Rabbit anti-Homo sapiens (Human) HRAS Polyclonal antibody
Alternative Names
C BAS/HAS antibody; C HA RAS1 antibody; C-BAS/HAS antibody; c-H-ras antibody; C-HA-RAS1 antibody; CTLO antibody; GTPase HRas antibody; GTPase KRas antibody; GTPase NRas antibody; H ras antibody; H RASIDX antibody; H-Ras-1 antibody; H-RASIDX antibody; Ha-Ras antibody; HAMSV antibody; HRAS antibody; HRAS1 antibody; K ras antibody; K RAS2A antibody; K RAS2B antibody; K RAS4A antibody; K RAS4B antibody; K-RAS antibody; KRAS antibody; KRAS1 antibody; KRAS2 antibody; N-RAS antibody; N-terminally processed antibody; NRAS antibody; NRAS1 antibody; p21ras antibody; RASH_HUMAN antibody; RASH1 antibody; RASK2 antibody; Transforming protein p21 antibody; v Ha ras Harvey rat sarcoma viral oncogene homolog antibody; v Ki ras2 Kirsten rat sarcoma viral oncogene homolog antibody; v ras neuroblastoma RAS viral oncogene homolog antibody
Immunogen
Recombinant Human GTPase HRas protein (12-181AA)
Immunogen Species
Homo sapiens (Human)
Conjugate
Non-conjugated
The HRAS Antibody (Product code: CSB-PA010726LA01HU) is Non-conjugated. For HRAS Antibody with conjugates, please check the following table.
Available Conjugates
| Conjugate |
Product Code |
Product Name |
Application |
| HRP |
CSB-PA010726LB01HU |
HRAS Antibody, HRP conjugated |
ELISA |
| FITC |
CSB-PA010726LC01HU |
HRAS Antibody, FITC conjugated |
|
| Biotin |
CSB-PA010726LD01HU |
HRAS Antibody, Biotin conjugated |
ELISA |
Purification Method
>95%, Protein G purified
Concentration
It differs from different batches. Please contact us to confirm it.
Buffer
Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, PH 7.4
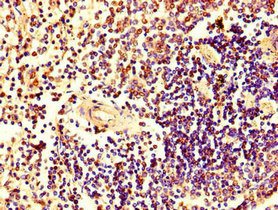
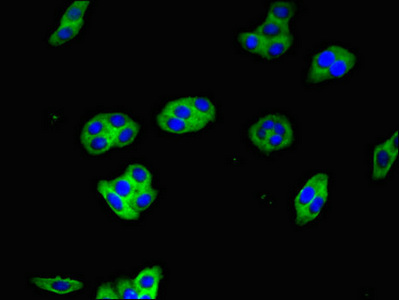
Tested Applications
ELISA, IHC, IF
Recommended Dilution
| Application |
Recommended Dilution |
| IHC |
1:20-1:200 |
| IF |
1:50-1:200 |
Storage
Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
Lead Time
Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
Usage
For Research Use Only. Not for use in diagnostic or therapeutic procedures.