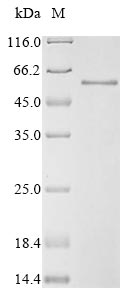
This Recombinant Human SMAD3 protein is specifically designed to support your transcription research. This full-length protein (1-425aa) is expressed in a Baculovirus expression system and is equipped with an N-terminal 10xHis-tag, facilitating effortless purification and detection. Our SMAD3 protein has a purity of greater than 85% as determined by SDS-PAGE, ensuring consistent performance and reliable results.
Dive into the complex functions of Mothers against decapentaplegic homolog 3 with our premium recombinant protein. Available in both liquid and lyophilized powder forms, our Recombinant Human SMAD3 protein offers the versatility and quality you need to advance your research endeavors and uncover new insights.