Human Cytomegalovirus
Human cytomegalovirus (CMV) has been infecting humans since the inception of the human species and currently affects the majority of the global population. HCMV is intimately related to various ailments, including tumors, circulatory system disorders, digestive system disorders, and idiopathic ophthalmic diseases.
1. What Is Human Cytomegalovirus?
Human cytomegalovirus (herpesvirus type 5, HHV-5/HCMV) is a double-stranded DNA β-herpesvirus belonging to the Herpesviridae family, the subfamily Betaherpesvirinae. HCMV is a common virus that infects 50 to 80 percent of people at some time during their lives but rarely causes obvious illness. Diseases associated with HHV-5 include mononucleosis and pneumonia. HCMV infection can become dormant for a while and may reactivate later. The virus is carried by people and is not associated with food, water, or animals.
2. Human Cytomegalovirus Structure and Genome
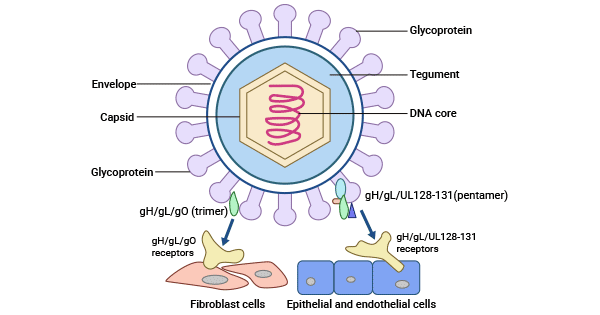
HCMV exhibits a distinctive three-layer structure comprising an outer lipid bilayer envelope, an inner icosahedral capsid enclosing the genome, and a middle pleomorphic tegument compartment. The HCMV capsid mainly consists of four components: the major capsid protein (MCP/pUL86), the triplex dimer (TRI2/pUL85), the triplex monomer (TRI1/pUL46), and the smallest capsid protein (SCP/pUL48A).
The CMV genome is segmented into two unique sequences, unique long (UL) and unique short (US), which are flanked by terminal and internal repeated sequences and arrayed as TRL-UL-IRL-IRS-US-TRS.
Figure 1. The structure of HCMV
This picture is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9784992/
This Table information is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC521840/ and https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7378892/
3. Human Cytomegalovirus Life Cycle
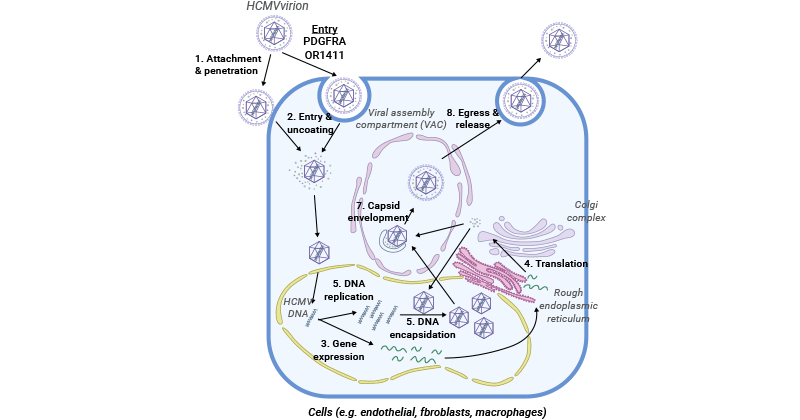
HCMV life cycle includes entry, viral replication and gene expression, viral assembly, encapsidation, and maturation, and finally release from the host cell [13].
Figure 2. The HCMV life cycle
This picture is cited from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7298070/
3.1 Entry
HCMV infects host cells like fibroblasts through membrane fusion that involves a trimer complex composed of gH/gL/gO (UL75/UL115/UL74) interacting with platelet-derived growth factor receptor α (PDGFRα) [1,2]. TGFβRIII and neuregulin-2 (NRG2) are additional receptors for trimers, facilitating viral entry in fibroblasts [1,4]. The gM/gN (UL100/UL73) surface dimer and the gB (UL55) trimer play essential roles in viral entry, with gM/gN facilitating adhesion through interaction with heparin sulfate proteoglycans on the cell surface and the gB trimer acting as a proximal mediator during membrane fusion.
HCMV can also enter host cells like epithelial and endothelial cells via low-pH endocytosis mediated by the interaction between a pentamer envelope glycoprotein complex composed of gH/gL/UL128/UL130/UL131 and neuropilin-2 (Nrp2) [1,3]. CD147, CD46, and OR14I1 have been also demonstrated to participate in the epithelial entry [1,4-6].
3.2 Viral Replication and Gene Expression
After passing through the host cell membrane, HCMV’s tegumented capsid is transported along the microtubules to the nuclear pore through the interaction between viral tegument proteins and the host microtubule machinery. The viral genome is released into the host nucleus through the nuclear pore. Some tegument proteins such as pp65 (pUL83) and pp71 (pUL82) dissociate with the viral capsid and are independently transported into the nucleus. pp65 plays a role in viral replication, while pp71 primarily influences gene expression.
The incoming viral genome serves as a template both for de novo transcription for viral protein synthesis and the generation of progeny genomes. In the host nucleus, the viral genome is circularized and forms a concatemeric DNA. pp71 attaches to the death domain-associated protein (Daxx), which binds to HDACs to inhibit transcription, thus leading to the degradation of Daxx and activating the expression of the immediate early (IE) genes [7]. The proteins of IE-encoding products interacting with cytokines further drive the expression of early (E) genes and late (L) genes [8]. E proteins are mainly responsible for viral transcription and viral DNA synthesis, while L proteins are primarily viral structural proteins.
HCMV encodes six core viral replication proteins, including UL54 (DNA polymerase), UL44 (DNA polymerase processivity factor), UL70 (primase), UL105 (helicase), UL102 (primase-associated factor), and UL57 (single-stranded DNA-binding protein).
3.3 Assembly, Maturation, and Release
The expression of L genes initiates viral capsid assembly in the nucleus. After capsid maturation, a terminase complex pUL51/pUL52/pUL56/pUL77/pUL89/pUL93 binds to the concatemeric viral DNA and cleaves it into unit-length genomes for subsequent package into procapsid to form the nucleocapsid [9-11].
After viral DNA replication and packaging of newly formed capsids, capsids containing the genome must move from replication compartments (RC) and traverse the nuclear envelope in an envelopment-deenvelopment-reenvelopment manner. This involves envelopment at the inner nuclear membrane (INM) and de-envelopment at the outer nuclear membrane (ONM), as well as additional tegumentation and final envelopment upon the capsid transported to the virion assembly compartment (vAC) in the cytoplasm [12]. The mature virion particles are then released from the cell by budding.
4. Human Cytomegalovirus Infection
HCMV can infect various cell types, including epithelial and endothelial cells, fibroblasts, and myeloid cells. HCMV infection can result in either lytic or latent infection, depending on the control of the viral major immediate early promoter (MIEP). During lytic infection, the MIEP, responsible for the expression of crucial lytic cycle genes (IE genes), is linked with transcriptional activation markers [17]. The lytic (productive) life cycle involves a temporal cascade of IE, E, and L gene expression, culminating in the production of progeny virions [18].
HCMV can establish lifelong latency following initial lytic infection, which facilitates its escape from immune clearance [16]. Latent infection has so far been found only in cells of cells of the early myeloid lineage, including CD34+ hematopoietic stem cells (HSCs), monocyte progenitors, and blood monocytes. In latently infected cells, when there is no production of new virions, the MIEP is related to inhibitory chromatin marks, and the viral genome is repressed [15]. Reactivation arises from CD34+ hematopoietic progenitor cells (HPCs)- and CD14+ monocytes-derived latency, frequently following blood transfusion and organ transplantation [14,15].
HCMV infection usually causes mild or no obvious symptoms in healthy individuals but is highly pathogenic in congenitally infected infants and immunocompromised patients such as transplant recipients.
5. Human Cytomegalovirus Transmission
The primary infection of HCMV is typically transmitted through intrauterine, breast milk, and exposure to contaminations such as saliva or genital secretions. HCMV is spread from an infected person by mucosal contact in the following ways:
-
From direct contact with saliva or urine, especially from babies and young children
-
Through sexual contact
-
From breast milk to nursing infants
If a pregnant woman is infected, the fetus may acquire the infection during the pregnancy, or the baby may acquire the infection during delivery.
-
Through transplanted organs and blood transfusions
People who have received an organ transplant are particularly susceptible to HCMV infection because they are given drugs that suppress the immune system to prevent rejection of the transplant.
6. Immune Response to Human Cytomegalovirus Infection
During HCMV primary infection, ample antibodies against multiple HCMV proteins are inductively produced in the host cells. These proteins include structural tegument proteins, envelope glycoproteins, and nonstructural proteins. Studies have indicated that humoral immunity is essential in limiting viral propagation and may play a role in reducing the clinical manifestations of the disease [19].
After entry into the targeted cells, numerous HCMV gene products are involved in the blockade of natural killer (NK) cell-mediated recognition to avoid immune surveillance and subsequent elimination. There are approximately 12 HCMV gene products known to modulate NK cell regulation, including US20, UL16, UL17, UL18, UL40, UL43, UL140, UL83, UL141-UL144, and UL148. UL16-, UL17-, UL40-, UL140-, and UL142-encoding products mimic host HLA class I and contribute to the down-regulation of NK cell activity. Additionally, the HCMV microRNA miR-UL122 functions to inhibit host MICB surface expression [20].
References
[1] Nguyen, C. C., and Kamil, J. P. (2018). Pathogen at the gates: human cytomegalovirus entry and cell tropism [J]. Viruses 10:704.
[2] Wu, Y., Prager, A., et al. (2017). Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry [J]. PLoS Pathog. 13:e1006281.
[3] Martinez-Martin, N., Marcandalli, J., et al. (2018). An unbiased screen for human cytomegalovirus identifies Neuropilin-2 as a central viral receptor [J]. Cell 174:28.
[4] Vanarsdall, A. L., Pritchard, et al. (2018). CD147 promotes entry of pentamer-expressing human cytomegalovirus into epithelial and endothelial cells [J]. mBio 9:e00781-18.
[5] Stein, K. R., Gardner, T. J., et al. (2019). CD46 facilitates entry and dissemination of human cytomegalovirus [J]. Nat. Commun. 10:2699.
[6] iaofei, E., Meraner, P., et al. (2019). OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism [J]. Proc. Natl. Acad. Sci. U.S.A. 116, 7043–7052.
[7] Saffert, R. T., and Kalejta, R. F. (2006). Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression [J]. J. Virol. 80, 3863–3871.
[8] Adamson, C. S., and Nevels, M. M. (2020). Bright and early: inhibiting human cytomegalovirus by targeting major immediate-early gene expression or protein function [J]. Viruses 12:110.
[9] Köppen-Rung, P., Dittmer, A., and Bogner, E. (2016). Intracellular Distribution of Capsid-Associated pUL77 of human cytomegalovirus and interactions with packaging proteins and pUL93 [J]. J. Virol. 90, 5876–5885.
[10] DeRussy, B. M., and Tandon, R. (2015). Human Cytomegalovirus pUL93 is required for viral genome cleavage and packaging [J]. J. Virol. 89, 12221–12225.
[11] Borst, E. M., Bauerfeind, R., et al. (2016). The essential human cytomegalovirus proteins pUL77 and pUL93 are structural components necessary for viral genome encapsidation [J].
[12] Sanchez, V., Greis, K. D., Sztul, E., and Britt, W. J. (2000). Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly [J]. J. Virol. 74, 975–986.
[13] Jean Beltran PM, Cristea IM. The life cycle and pathogenesis of human cytomegalovirus infection: lessons from proteomics. Expert Rev Proteomics [J]. 2014 Dec;11(6):697-711.
[14] Zhao, X.-Y., Luo, X.-Y., et al. (2017). Recipient-donor KIR ligand matching prevents CMV reactivation post-haploidentical T cell-replete transplantation [J]. Br. J. Haematol. 177, 766–781.
[15] Sinclair J., Poole E. Human cytomegalovirus latency and reactivation in and beyond the myeloid lineage [J]. Future Virol. 2014;6:7.
[16] Poole E., Wills M., Sinclair J. Human Cytomegalovirus Latency: Targeting Differences in the Latently Infected Cell with a view to Clearing Latent Infection [J]. New J. Sci. 2014;2014:10.
[17] Poole E, Lau J, et al. The Human Cytomegalovirus Latency-Associated Gene Product Latency Unique Natural Antigen Regulates Latent Gene Expression [J]. Viruses. 2023 Sep 4;15(9):1875.
[18] Wang YQ, Zhao XY. Human Cytomegalovirus Primary Infection and Reactivation: Insights From Virion-Carried Molecules [J]. Front Microbiol. 2020 Jul 14;11:1511.
[19] Gerna G, Sarasini A, Patrone M, et al. Human cytomegalovirus serum neutralizing antibodies block virus infection of endothelial/epithelial cells, but not fibroblasts, early during primary infection [J]. J Gen Virol. 2008;89(Pt 4):853–865.
[20] Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miRUL112 acts synergistically with a cellular microRNA to escape immune elimination [J]. Nat Immunol. (2010) 11:806–813.