What is Mitophagy?
Mitophagy, one of selective autophagy, specifically gets rid of damaged or extra mitochondria. Simply speaking, mitophagy means the degradation of dysfunctional or superfluous mitochondria.
The Function of Mitophagy
A mitochondrion is a place where eukaryotic cells undergo bio-oxidation and energy conversion. It can provide most of the energy for life activities through oxidative phosphorylation, so it is known as "energy factory". However, mitochondria also produce reactive oxygen species (ROS) while generating ATP to supply energy. Abnormal accumulation of ROS can cause mitochondrial damage such as mitochondrial DNA (mt DNA), lipids and proteins, which in turn make mitochondria vulnerable to producing ROS productions.
Mitochondria damage disrupts mitochondrial function, leading to the accumulation of defective organelles, and further threatening the survival of cell and tissue. Besides, dysfunctional mitochondria have been demonstrated to involve in aging, diabetes, and neurodegenerative diseases. Therefore, timely elimination of partial mitochondria in cells plays an important role in balancing the quality and quantity of the mitochondria, thus maintaining cell homeostasis. Eukaryotic cells primarily clear damaged mitochondria through mitophagy, although fraction mitochondria are directly engulfed by autophagy.
The Universal Process of Mitophagy
Usually, mitophagy includes three consecutive steps: the sequestration of a double-membrane vesicle enclosing a complete mitochondrion, the formation of autophagosome, and the fusion with a lysosome for ultimate degradation.
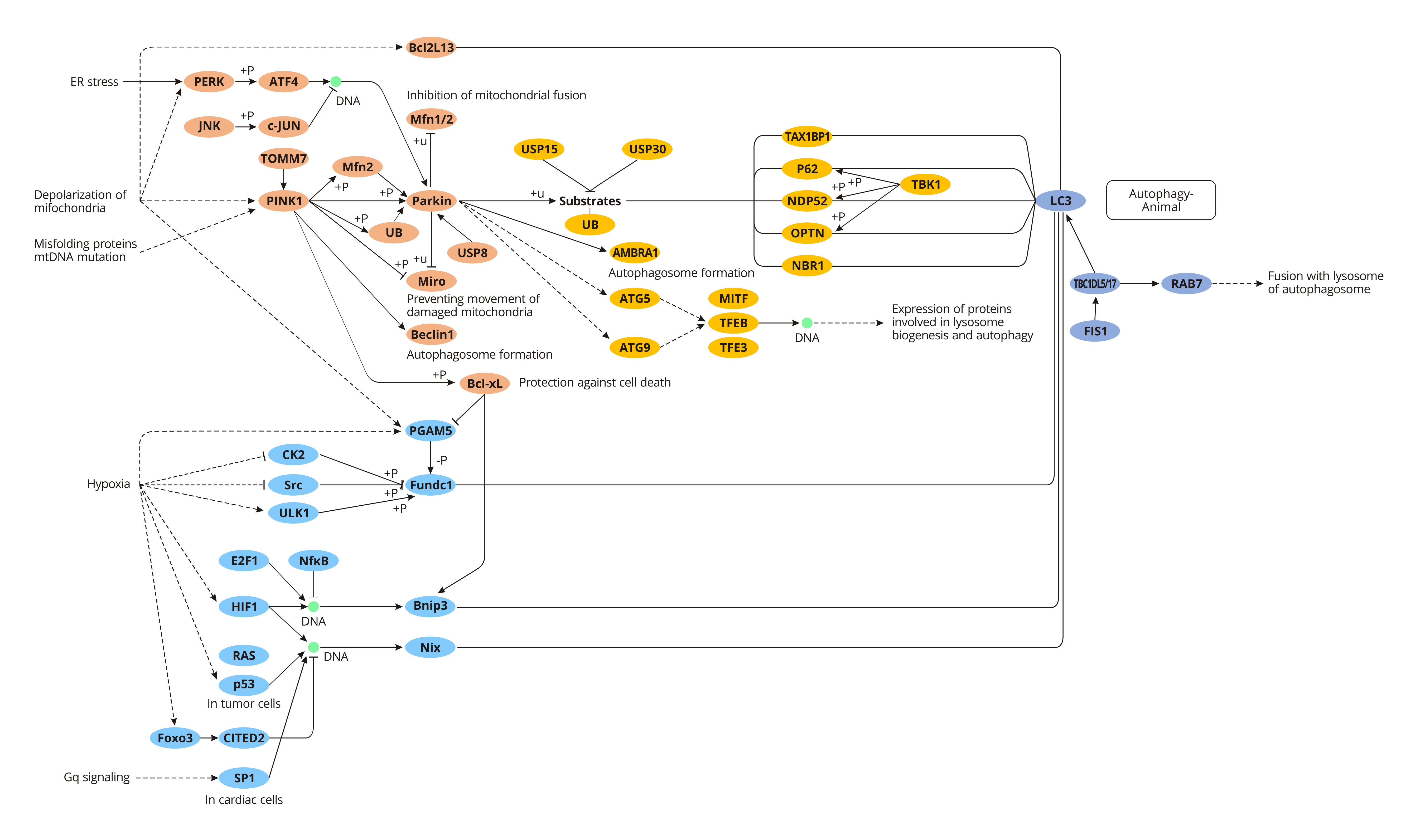
Regulation Mechanism of Mitophagy
Mitophagy exists in yeast, plants, and animals. Its induction and regulation mechanisms are either similar and different. Here primarily introduce the regulation mechanism of mitophagy in yeast and mammals in detail.
In Yeast
In yeast, mitochondrial autophagy is mainly mediated by ATG32, which is the mitochondrial outer membrane protein of yeast, and its N-terminal & c-terminal are exposed to cytoplasm and mitochondrial stroma respectively. When mitochondria are damaged, ATG32 phosphorylation on mitochondrial membrane mediates the connection with ATG11 and locates mitochondria at phagophore assembly site (PAS). At the same time, ATG32 connects with ATG8 through its LIR domain to initiate the formation of autophagosome, thus inducing the degradation of the mitochondrial autophagy pathway.
In mammals
Parkin/ pink1-mediated Mitophagy
Parkin is an E3 ubiquitin-protein ligase, which is highly expressed in the brain, skeletal muscle, cardiac muscle, liver, etc, and is free in the cytoplasm. Under normal physiological condition, PINK1 (PTEN-induced putative kinase 1) combines with TOM (mitochondrial outer membrane translocase) to form a compound, entering into the mitochondrial intermembrane space, where the complex interacts with TIM (inner membrane translocase). And the complex is quickly degraded by PARL to ensure the low-level PINK1 and normal mitophagy.
However, when mitochondria are damaged or dysfunctional, the membrane potential of mitochondria is weakened, and PINK1 no longer transports to the inner membrane and binds to TOM, but locates in the outer membrane of mitochondria and recruits Parkin to mitochondria from the cytoplasm. The activation of Parkin by PINK1 leads to cascade ubiquitination of downstream mitochondrial proteins, such as MARF, Mfn1, Mfn2 and voltage-dependent cation channel protein 1 (VDAC1), inhibiting the fluidity of damaged mitochondria and their fusion with normal mitochondria, thereby promoting the specific recognition and wrapping of damaged mitochondria by autophagosomes, and eventually removing the damaged mitochondria.
FUNDC1-mediated Mitophagy
FUNDC1, a new receptor molecule discovered in 2012 that is involved in regulating mitochondrial autophagy in mammalian cells, is highly conserved in most mammals. FUNDC1, a transmembrane protein, locates on the outer membrane of mitochondria and has 3 alpha-helices. The N terminal of FUNDC1 contains a scope LIR (lc3-interaction region) directly bound to LC3. Under normal physiological conditions, protein kinase Src inhibits the action of FUNDC1 and LC3 phosphorylates LIR, thereby down-regulating mitochondrial autophagy activity. On the contrary, under hypoxic environment, the activity of protein kinase Scr is inhibited, and the phosphorylation level of FUNDC1 is reduced, leading to enhanced binding of FUNDC1 to LC3, which in turn activates downstream mitophagy. The interaction between FUNDC1 and LC3 is inhibited when LIR is absent or mutated, which will further inhibit mitophagy.
Nix-mediated Mitophagy
Nix, also known as BCL2 ligin (BNIP3L), is a kind of mitochondrial membrane surface binding protein, which can directly interact with LC3 and GABARAP to induce autophagy degradation of mitochondria. In the maturation process of mammalian erythrocytes, nix-mediated mitochondrial autophagy plays a crucial role in the removal of mitochondria. Besides, studies have also found that there are similar structural domains and cellular functions between Nix protein and ATG32 in yeast mitochondrial autophagy. For example, the TM structure at carboxyl-terminal is responsible for connecting with the mitochondrial membrane, and LIR domain can connect with ATG8 family proteins.
Two Paradigms of Mitophagy
Mature red blood cells (RBCs) or erythrocytes have no nuclei or mitochondria but are rich in hemoglobin. RBCs use hemoglobin to transport oxygen that inhaled alveoli to the tissues in exchange for the RBCs to expel some of the CO2 produced by tissue metabolism through the alveoli. For the RBC to function as an oxygen transporter, the precursor reticular cell must clear most organelles to space more room for hemoglobin to mature into an RBC. Indeed, erythrocyte precursor reticular cells degrade mitochondria through mitophagy to achieve this goal.
In most cases, mammalian mitochondria all come from their mothers, although a few people have been recently observed to have small amounts of paternal mitochondria. This is due to the elimination of sperm-derived mitochondria after fertilization. The clearance of paternal mitochondria by mitophagy confers the maternal inheritance of mitochondria. However, there is no clear evidence of how this process occurs and why it does like this.
Diseases and Mitophagy
The main pathological changes in Parkinson's Disease (PD) are the progressive loss of dopaminergic neurons in the substantia nigra, which leads to decreased dopamine synthesis in the brain. As dopamine is an excitatory neurotransmitter, its decreased expression can lead to the imbalance of excitatory and inhibitory neurotransmitters. And the imbalance could bring symptoms in body such as static tremor, increased muscle tension and gait disorder. Abnormal mitochondrial autophagy is closely related to the occurrence of neurodegenerative diseases such as PD.
Mitophagy is also involved in cell aging. Mitochondria are a source of continuous ROS (reactive oxygen species). These ROS lead to mitochondrial DNA mutations, and the weak repairability of mitochondrial DNA causes the gradual accumulation of these mutations. Mutations in mitochondrial DNA may weaken the synthesis of proteins critical to oxidative phosphorylation, resulting in eventual nuclear cell death. As the body ages, the autophagy mechanism of cells gradually weakens, making damaged mitochondria gradually accumulate, and eventually aggravate aging.





