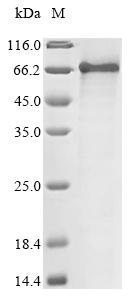
The process of synthesizing the recombinant bovine ALB protein includes transfecting yeast cells with a DNA expression vector containing the gene for the ALB protein. Subsequently, the cells are cultured to induce the expression of the intended protein. The recombinant bovine ALB protein is then collected and purified from the cell lysate through affinity purification. It exhibits a purity exceeding 85%, as verified by SDS-PAGE.
Albumin, a crucial protein in the human body, plays various essential roles due to its unique properties [2]. It acts as a physiological buffer, maintaining colloid oncotic pressure in healthy individuals [1]. Human serum albumin (HSA), the most abundant protein in blood circulation, exhibits extraordinary ligand binding capacity, making it a transporter for a wide range of substances [3][4]. Albumin is involved in critical processes such as endocytosis, which is particularly relevant in neurons [5].
Albumin interacts with various molecules and undergoes modifications, as observed in the increased clearance of glycated albumin by proximal tubule cells [6]. The evolution of albumin can be traced back to early vertebrates, indicating its fundamental presence in mammalian plasma [7]. The production of serum albumin increases significantly after birth, becoming the predominant protein in adult serum [8].
References:
[1] S. Haque, F. Kabir, & K. Haque, Albumin infusion therapy in critical patients, Bangladesh Critical Care Journal, vol. 6, no. 2, p. 88-91, 2018. https://doi.org/10.3329/bccj.v6i2.38584
[2] R. García-Martinez, P. Caraceni, M. Bernardi, P. Ginès, V. Arroyo, & R. Jalan, Albumin: pathophysiologic basis of its role in the treatment of cirrhosis and its complications, Hepatology, vol. 58, no. 5, p. 1836-1846, 2013. https://doi.org/10.1002/hep.26338
[3] K. Neelofar, Z. Arif, J. Ahmad, & K. Alam, Non-enzymatic glucosylation induced neo-epitopes on human serum albumin: a concentration based study, Plos One, vol. 12, no. 2, p. e0172074, 2017. https://doi.org/10.1371/journal.pone.0172074
[4] K. Majorek, P. Porebski, A. Dayal, M. Zimmerman, K. Jablonska, A. Stewartet al., Structural and immunologic characterization of bovine, horse, and rabbit serum albumins, Molecular Immunology, vol. 52, no. 3-4, p. 174-182, 2012. https://doi.org/10.1016/j.molimm.2012.05.011
[5] L. Rasgado, A. Urbieta, & J. Jiménez, Affected albumin endocytosis as a new neurotoxicity mechanism of amyloid beta, Aims Neuroscience, vol. 7, no. 3, p. 344-359, 2020. https://doi.org/10.3934/neuroscience.2020021
[6] M. Wagner, J. Myslinski, S. Pratap, B. Flores, G. Rhodes, S. Campos-Bilderbacket al., Mechanism of increased clearance of glycated albumin by proximal tubule cells, Ajp Renal Physiology, vol. 310, no. 10, p. F1089-F1102, 2016. https://doi.org/10.1152/ajprenal.00605.2015
[7] A. Bujacz, Structures of bovine, equine and leporine serum albumin, Acta Crystallographica Section D Biological Crystallography, vol. 68, no. 10, p. 1278-1289, 2012. https://doi.org/10.1107/s0907444912027047
[8] L. Jagodzinski, T. Sargent, M. Yang, C. Glackin, & J. Bonner, Sequence homology between rnas encoding rat alpha-fetoprotein and rat serum albumin., Proceedings of the National Academy of Sciences, vol. 78, no. 6, p. 3521-3525, 1981. https://doi.org/10.1073/pnas.78.6.3521