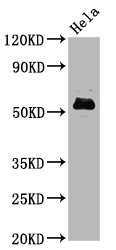
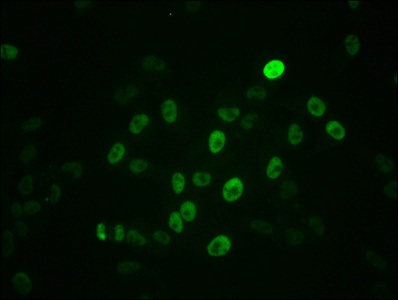
In the development of the phospho-MYC (S62) recombinant monoclonal antibody, the initial phase comprises the retrieval of genes responsible for coding the MYC antibody. These genes are acquired from rabbits that have been previously exposed to a synthesized peptide derived from the human MYC protein phosphorylated at S62. Subsequently, these antibody genes are seamlessly integrated into specialized expression vectors. Following this genetic modification, the vectors are introduced into host suspension cells, which are carefully cultured to stimulate the expression and secretion of antibodies. Following this cultivation phase, the phospho-MYC (S62) recombinant monoclonal antibody is subjected to a thorough purification process utilizing affinity chromatography techniques, effectively separating the antibody from the surrounding cell culture supernatant. Ultimately, the functionality of the antibody is comprehensively evaluated through a diverse array of assays, including ELISA, WB, and IF tests, unequivocally confirming its capacity to interact with the human MYC protein phosphorylated at S62.
Phosphorylation of MYC at S62 is a crucial regulatory mechanism that modulates MYC's transcriptional activity and function. Dysregulation of this phosphorylation event can have significant implications for cancer development and progression, making MYC an important target for cancer research and therapy.