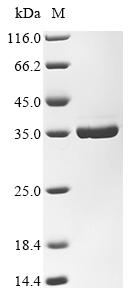
The expression of recombinant Escherichia phage RB69 (Bacteriophage RB69) DNA-directed DNA polymerase (Gp43) protein involves transfecting e.coli cells with a DNA expression vector carrying the gene for the protein of interest (103-340aa). After transfection, the cells are cultured to produce the desired protein. A N-terminal 10xHis tag and C-terminal Myc tag is fused to the protein. The recombinant Escherichia phage RB69 (Bacteriophage RB69) DNA-directed DNA polymerase (Gp43) protein is collected and purified from the cell lysate through affinity purification, attaining a purity greater than 90%, as determined by SDS-PAGE.