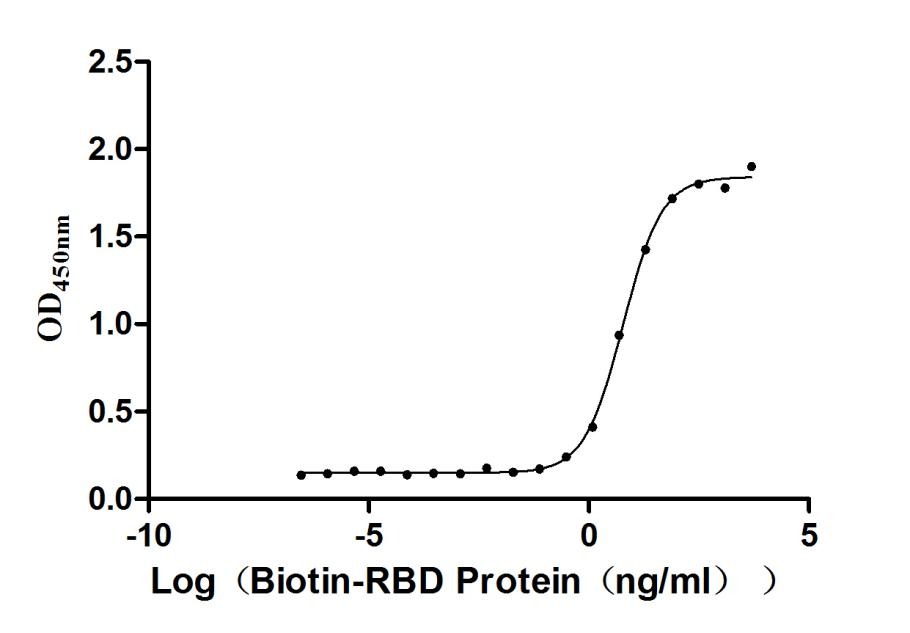
Recombinant severe acute respiratory syndrome coronavirus 2 Spike glycoprotein (S) production involves several steps starting with the isolation of the target gene that corresponds to the 319-541aa of the SARS-CoV-2 S protein. The gene is fused with a C-terminal mFC-Avi-tag gene and then cloned into an expression vector and introduced into mammalian cells via transformation. The mammalian cells express the recombinant protein, which is harvested from the cell lysate. The protein is purified using affinity chromatography. The final step involves validating the protein's purity and functionality. Its purity is determined through SDS-PAGE and SEC-HPLC, reaching over 90% and 95%, respectively. Its endotoxin content is less than 1.0 EU/ug as determined by the LAL method. This biotinylated recombinant SARS-CoV-2 S protein has been validated as an active protein in a functional ELISA.
The S protein of SARS-CoV-2 is a crucial component of the virus responsible for mediating host-cell entry. It is a type-1 transmembrane glycoprotein composed of two subunits, S1 and S2, responsible for attachment and entry of the virus, respectively [1][2]. The S protein is highly glycosylated and projects from the viral surface, facilitating attachment to the ACE2 receptor on host cells [3]. This protein is the main target for vaccine development and clinical diagnosis due to its antigenicity and role in virus entry [4][5]. Additionally, the S protein plays a key role in syncytia formation and virus entry by associating with the ACE2 receptor [5].
References:
[1] Y. Huang, C. Yang, X. Xu, W. Xü, & S. Liu, Structural and functional properties of sars-cov-2 spike protein: potential antivirus drug development for covid-19, Acta Pharmacologica Sinica, vol. 41, no. 9, p. 1141-1149, 2020. https://doi.org/10.1038/s41401-020-0485-4
[2] M. Hussain, N. Jabeen, A. Amanullah, B. Aa, B. Aziz, S. Shabbiret al., Structural basis of sars-cov-2 spike protein priming by tmprss2,, 2020. https://doi.org/10.1101/2020.04.21.052639
[3] U. Shekhawat and A. Chowdhury, Computational and comparative investigation of hydrophobic profile of spike protein of sars-cov-2 and sars-cov, Journal of Biological Physics, vol. 48, no. 4, p. 399-414, 2022. https://doi.org/10.1007/s10867-022-09615-x
[4] J. Zhou, J. Sun, G. Lu, W. Wang, & W. Lin, Sars-cov-2 spike protein evolution may cause difficulties for vaccine,, 2020. https://doi.org/10.21203/rs.3.rs-80010/v1
[5] D. Li, Y. Liu, Y. Lu, S. Gao, & L. Zhang, Palmitoylation of sars‐cov‐2 s protein is critical for s‐mediated syncytia formation and virus entry, Journal of Medical Virology, vol. 94, no. 1, p. 342-348, 2021. https://doi.org/10.1002/jmv.27339