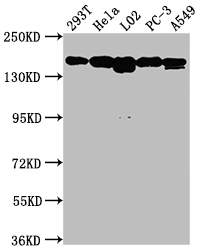
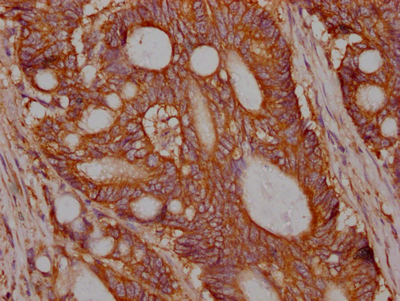
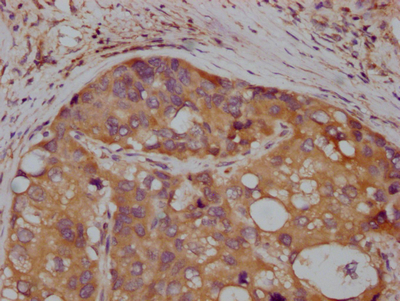
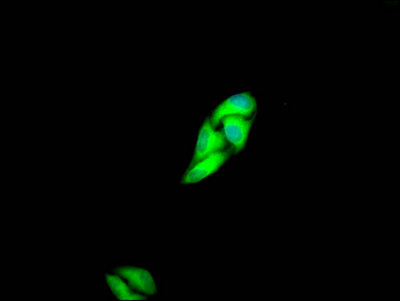
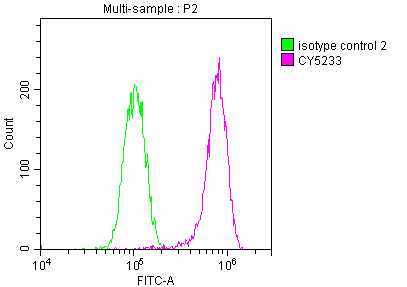
The MET recombinant monoclonal antibody is developed through protein and DNA recombinant technologies. Initially, a synthesized peptide derived from human MET protein is used to immunize mice. Spleen cells are extracted under sterile conditions from the immunized mice and cDNA synthesized by RNA reverse transcription is used as the PCR template for amplifying the MET antibody gene. Subsequently, the MET antibody gene is introduced into a vector and transfected into host cells for culture. Finally, the MET recombinant monoclonal antibody is purified from the cell culture supernatant using affinity chromatography. It has undergone rigorous verification and can be employed for detecting human MET protein in five experiments, including ELISA, WB, IHC, IF, and FC.
The MET protein, also known as hepatocyte growth factor receptor (HGFR), is a transmembrane receptor tyrosine kinase that is activated by binding of its ligand HGF. HGFR-HGF interaction activates downstream signaling pathways, including the RAS-MAPK pathway, PI3K-AKT pathway, and STAT3 pathway, among others. These pathways ultimately lead to cellular responses such as increased cell proliferation, survival, and migration. The dysregulation of MET has been implicated in various human diseases, including cancer.