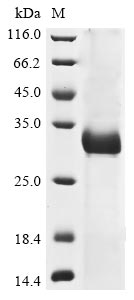
Recombinant Human coronavirus OC43 Spike glycoprotein (S) is produced in E. coli and spans the 318-624 amino acid region. This partial construct includes a C-terminal 6xHis-tag, which appears to make purification more straightforward. The protein shows a purity level greater than 85% as determined by SDS-PAGE, making it suitable for various research applications. It is intended for research use only and is not for diagnostic or therapeutic purposes.
The Spike glycoprotein of Human coronavirus OC43 plays a crucial role in viral entry into host cells, enabling attachment and fusion processes. It represents a key target for studying viral pathogenesis and immune response mechanisms. Understanding the structure and function of this protein may be essential in virology research, particularly in the context of coronavirus-related studies.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
The E. coli system lacks eukaryotic post-translational modifications (PTMs). While the His tag improves solubility, E. coli struggles to correctly fold eukaryotic membrane-associated proteins like the S protein—critical for functions such as receptor binding or interaction with other viral components. No data confirms native secondary/tertiary structure (e.g., circular dichroism for disulfide bonds, thermal shift assays for stability). E. coli may misfold the fragment, leading to aggregation or non-functional conformations. The fragment lacks full-length S context (e.g., receptor-binding domain [RBD], fusion machinery), so it cannot recapitulate native interactions (e.g., with host receptors like CEACAM1 for OC43). Bioactivity (e.g., ligand binding, oligomerization) is untested and unlikely to mirror wild-type S.
1. Antibody Development and Characterization
This recombinant S fragment can serve as an immunogen for generating antibodies, but antibody specificity must be validated against native S—E. coli-expressed protein may present non-native epitopes, leading to cross-reactivity with irrelevant targets. The His tag simplifies purification/immobilization for ELISA, but antibodies may not recognize the fragment in its native context (e.g., on viral particles).
2. Protein-Protein Interaction Studies
Pull-down assays using the His tag can identify interactors, but results depend on correct folding—E. coli-expressed fragments often misfold, causing false positives/negatives. Identified partners must be validated via co-IP or functional assays (e.g., replicon assays) to rule out artifacts. The partial length restricts insights to interactions within this domain, not full-length S biology.
3. Structural and Biochemical Characterization
This fragment supports preliminary biophysical studies (e.g., CD for secondary structure, DLS for stability) but cannot inform native S architecture—E. coli-expressed protein may lack correct disulfide bonds or domain folding. Structural conclusions (e.g., oligomerization) must be contextualized by folding limitations.
4. ELISA-Based Binding Assays
The His tag enables immobilization for ELISA, but misfolding may alter binding—results are qualitative at best. Quantitative affinity measurements require a confirmed native structure, as the fragment’s conformation may not support native-like interactions.
Final Recommendation & Action Plan
This E. coli-expressed HCoV-OC43 S fragment has limited utility without rigorous validation first, confirming folding via CD spectroscopy and thermal shift assays to rule out misfolding; second, testing bioactivity (e.g., binding to known ligands or host receptors) to ensure native-like function. Optimize expression (e.g., co-express chaperones, use low-temperature induction) to improve solubility. For antibody development, validate specificity against native S; for interactions/ELISA, use tag cleavage or orthogonal methods (e.g., SPR) to reduce artifacts. If folding/bioactivity fails, use a eukaryotic system (e.g., mammalian/insect cells) to ensure native structure—this fragment alone is insufficient for reliable downstream applications without validation.