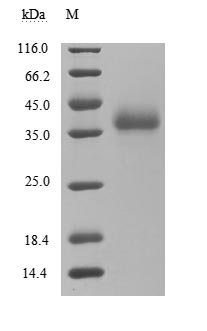
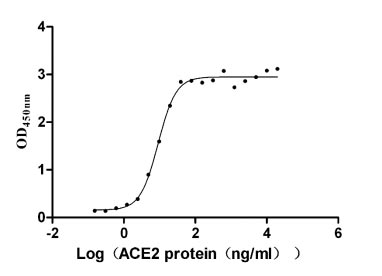
This recombinant protein corresponds to the receptor-binding domain (RBD, amino acids 306-527) of the SARS-CoV spike glycoprotein (S), expressed in mammalian cells with dual tags: an N-terminal 10×His tag and a C-terminal Myc tag for enhanced detection and purification. It demonstrates high purity (>95% by SDS-PAGE) and low endotoxin levels (<1.0 EU/μg, LAL method), ensuring suitability for sensitive microbiological assays. Functional validation via ELISA confirms its binding activity to both Paguma larvata ACE2 (CSB-MP684964PAL) and human ACE2 (CSB-MP866317HU), with EC50 of 5.056–7.559 ng/mL and 7.941–10.49 ng/mL, respectively, highlighting its utility in studying cross-species viral entry mechanisms and host receptor interactions. Provided as a lyophilized powder, this recombinant SARS-CoV S protein retains structural integrity and is optimized for reconstitution in downstream applications. Its mammalian expression ensures proper glycosylation and folding, making it a critical tool for SARS-CoV research, vaccine development, and therapeutic antibody screening in microbiology.
The spike (S) protein of SARS-CoV, a member of the coronavirus family, is a critical determinant of the virus's ability to infect host cells. This glycoprotein is integral to the virus's life cycle as it facilitates the initial attachment of the virus to host cells via binding to the ACE2 receptor. The S protein consists of two main subunits: the S1 subunit and the S2 subunit. The S1 subunit is primarily responsible for receptor recognition, while the S2 subunit plays a pivotal role in membrane fusion and the entry of the viral particle into the host cell [1][2].
The process of viral entry is dependent on several biochemical steps that include proteolytic cleavage of the S protein, which is essential for its fusion capability. In particular, cleavage at the S1/S2 site by host proteases such as the TMPRSS2 is crucial. This cleavage event activates the S protein, allowing it to facilitate membrane fusion between the viral and host cell membranes. Alternatively, in some cellular environments, other proteases such as Cathepsin B/L can also assist in this activation step [3][4][5]. Such proteolytic processing not only primes the S protein but also enhances the infectivity of the virus, underscoring the role of host factors in SARS-CoV and its related strains, including SARS-CoV-2 [4][6]. Moreover, the S protein is not only critical for the entry of the virus but also serves as a major target for vaccine development due to its immunogenic properties [1][2][7].
References:
[1] F. Cito, L. Amato, et al. A covid-19 hotspot area: activities and epidemiological findings, Microorganisms. vol. 8, no. 11, p. 1711, 2020. https://doi.org/10.3390/microorganisms8111711
[2] M. Medina-Enríquez, S. López‐León, J. Carlos-Escalante, Z. Aponte‐Torres, A. Cuapio, & T. Wegman‐Ostrosky. Ace2: the molecular doorway to sars-cov-2. Cell & Bioscience, vol. 10, no. 1, 2020. https://doi.org/10.1186/s13578-020-00519-8
[3] M. Hoffmann, H. Kleine‐Weber, & S. Pöhlmann. A multibasic cleavage site in the spike protein of sars-cov-2 is essential for infection of human lung cells. Molecular Cell, vol. 78, no. 4, p. 779-784.e5, 2020. https://doi.org/10.1016/j.molcel.2020.04.022
[4] M. Hoffmann, H. Kleine‐Weber, et al. Sars-cov-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell, vol. 181, no. 2, p. 271-280.e8, 2020. https://doi.org/10.1016/j.cell.2020.02.052
[5] G. Sun, Y. Sui, et al. Structural basis of covalent inhibitory mechanism of tmprss2-related serine proteases by camostat. Journal of Virology, vol. 95, no. 19, 2021. https://doi.org/10.1128/jvi.00861-21
[6] Y. Park, J. Ahn, S. Hwang, K. Sung, J. Lim, & K. Kwack. Structural similarity analysis of the spike protein of sars-cov-2 and other sars-related coronaviruses. 2020. https://doi.org/10.20944/preprints202003.0409.v1
[7] S. Jangra, J. Vrieze, et al. Sterilizing immunity against sars-cov-2 infection in mice by a single-shot and modified imidazoquinoline tlr7/8 agonist-adjuvanted recombinant spike protein vaccine. 2020. https://doi.org/10.1101/2020.10.23.344085